Inferring time series chromatin states for promoter-enhancer pairs based on Hi-C data
- PMID: 33509077
- PMCID: PMC7841892
- DOI: 10.1186/s12864-021-07373-z
Inferring time series chromatin states for promoter-enhancer pairs based on Hi-C data
Abstract
Background: Co-localized combinations of histone modifications ("chromatin states") have been shown to correlate with promoter and enhancer activity. Changes in chromatin states over multiple time points ("chromatin state trajectories") have previously been analyzed at promoter and enhancers separately. With the advent of time series Hi-C data it is now possible to connect promoters and enhancers and to analyze chromatin state trajectories at promoter-enhancer pairs.
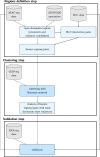
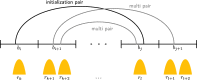
Results: We present TimelessFlex, a framework for investigating chromatin state trajectories at promoters and enhancers and at promoter-enhancer pairs based on Hi-C information. TimelessFlex extends our previous approach Timeless, a Bayesian network for clustering multiple histone modification data sets at promoter and enhancer feature regions. We utilize time series ATAC-seq data measuring open chromatin to define promoters and enhancer candidates. We developed an expectation-maximization algorithm to assign promoters and enhancers to each other based on Hi-C interactions and jointly cluster their feature regions into paired chromatin state trajectories. We find jointly clustered promoter-enhancer pairs showing the same activation patterns on both sides but with a stronger trend at the enhancer side. While the promoter side remains accessible across the time series, the enhancer side becomes dynamically more open towards the gene activation time point. Promoter cluster patterns show strong correlations with gene expression signals, whereas Hi-C signals get only slightly stronger towards activation. The code of the framework is available at https://github.com/henriettemiko/TimelessFlex .
Conclusions: TimelessFlex clusters time series histone modifications at promoter-enhancer pairs based on Hi-C and it can identify distinct chromatin states at promoter and enhancer feature regions and their changes over time.
Keywords: Chromatin immunoprecipitation; Differentiation; Enhancer; Gene regulation; Hi-C; Histone modifications.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- Libbrecht MW, Ay F, Hoffman MM, Gilbert DM, Bilmes JA, Noble WS. Joint annotation of chromatin state and chromatin conformation reveals relationships among domain types and identifies domains of cell type-specific expression. Genome Res. 2015;25(4):544–557. doi: 10.1101/gr.184341.114. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

