Arabidopsis cell wall composition determines disease resistance specificity and fitness
- PMID: 33509925
- PMCID: PMC7865177
- DOI: 10.1073/pnas.2010243118
Arabidopsis cell wall composition determines disease resistance specificity and fitness
Abstract
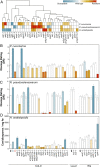
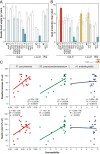
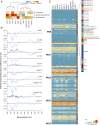
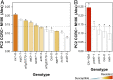
Plant cell walls are complex structures subject to dynamic remodeling in response to developmental and environmental cues and play essential functions in disease resistance responses. We tested the specific contribution of plant cell walls to immunity by determining the susceptibility of a set of Arabidopsis cell wall mutants (cwm) to pathogens with different parasitic styles: a vascular bacterium, a necrotrophic fungus, and a biotrophic oomycete. Remarkably, most cwm mutants tested (29/34; 85.3%) showed alterations in their resistance responses to at least one of these pathogens in comparison to wild-type plants, illustrating the relevance of wall composition in determining disease-resistance phenotypes. We found that the enhanced resistance of cwm plants to the necrotrophic and vascular pathogens negatively impacted cwm fitness traits, such as biomass and seed yield. Enhanced resistance of cwm plants is not only mediated by canonical immune pathways, like those modulated by phytohormones or microbe-associated molecular patterns, which are not deregulated in the cwm tested. Pectin-enriched wall fractions isolated from cwm plants triggered immune responses in wild-type plants, suggesting that wall-mediated defensive pathways might contribute to cwm resistance. Cell walls of cwm plants show a high diversity of composition alterations as revealed by glycome profiling that detect specific wall carbohydrate moieties. Mathematical analysis of glycome profiling data identified correlations between the amounts of specific wall carbohydrate moieties and disease resistance phenotypes of cwm plants. These data support the relevant and specific function of plant wall composition in plant immune response modulation and in balancing disease resistance/development trade-offs.
Keywords: cell wall; disease resistance; fitness; glycomics; immunity.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Couto D., Zipfel C., Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016). - PubMed
-
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. M., Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 (2012). - PubMed
-
- Shigenaga A. M., Berens M. L., Tsuda K., Argueso C. T., Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Biol. 38, 164–172 (2017). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

