3D Functional Genomics Screens Identify CREBBP as a Targetable Driver in Aggressive Triple-Negative Breast Cancer
- PMID: 33509944
- PMCID: PMC7611219
- DOI: 10.1158/0008-5472.CAN-20-1822
3D Functional Genomics Screens Identify CREBBP as a Targetable Driver in Aggressive Triple-Negative Breast Cancer
Abstract
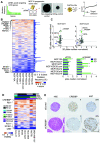
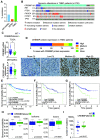
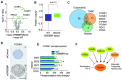
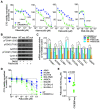
Triple-negative breast cancers (TNBC) are resistant to standard-of-care chemotherapy and lack known targetable driver gene alterations. Identification of novel drivers could aid the discovery of new treatment strategies for this hard-to-treat patient population, yet studies using high-throughput and accurate models to define the functions of driver genes in TNBC to date have been limited. Here, we employed unbiased functional genomics screening of the 200 most frequently mutated genes in breast cancer, using spheroid cultures to model in vivo-like conditions, and identified the histone acetyltransferase CREBBP as a novel tumor suppressor in TNBC. CREBBP protein expression in patient tumor samples was absent in 8% of TNBCs and at a high frequency in other tumors, including squamous lung cancer, where CREBBP-inactivating mutations are common. In TNBC, CREBBP alterations were associated with higher genomic heterogeneity and poorer patient survival and resulted in upregulation and dependency on a FOXM1 proliferative program. Targeting FOXM1-driven proliferation indirectly with clinical CDK4/6 inhibitors (CDK4/6i) selectively impaired growth in spheroids, cell line xenografts, and patient-derived models from multiple tumor types with CREBBP mutations or loss of protein expression. In conclusion, we have identified CREBBP as a novel driver in aggressive TNBC and identified an associated genetic vulnerability in tumor cells with alterations in CREBBP and provide a preclinical rationale for assessing CREBBP alterations as a biomarker of CDK4/6i response in a new patient population. SIGNIFICANCE: This study demonstrates that CREBBP genomic alterations drive aggressive TNBC, lung cancer, and lymphomas and may be selectively treated with clinical CDK4/6 inhibitors.
©2021 American Association for Cancer Research.
Conflict of interest statement
ANT has served on advisory boards for Pfizer. SPV is a Chief investigator on a number of clinical trials involving other compounds with Pfizer. LAM receives academic funding from Pfizer. No potential conflicts of interest were disclosed by the other authors.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

