Direct allele introgression into pure chicken breeds using Sire Dam Surrogate (SDS) mating
- PMID: 33510156
- PMCID: PMC7844028
- DOI: 10.1038/s41467-020-20812-x
Direct allele introgression into pure chicken breeds using Sire Dam Surrogate (SDS) mating
Abstract
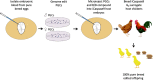
Poultry is the most abundant livestock species with over 60 billion chickens raised globally per year. The majority of chicken are produced from commercial flocks, however many indigenous chicken breeds play an important role in rural economies as they are well adapted to local environmental and scavenging conditions. The ability to make precise genetic changes in chicken will permit the validation of genetic variants responsible for climate adaptation and disease resilience, and the transfer of beneficial alleles between breeds. Here, we generate a novel inducibly sterile surrogate host chicken. Introducing donor genome edited primordial germ cells into the sterile male and female host embryos produces adult chicken carrying only exogenous germ cells. Subsequent direct mating of the surrogate hosts, Sire Dam Surrogate (SDS) mating, recreates the donor chicken breed carrying the edited allele in a single generation. We demonstrate the introgression and validation of two feather trait alleles, Dominant white and Frizzle into two pure chicken breeds using the SDS surrogate hosts.
Conflict of interest statement
The authors M.W. and M.J.M. are inventors on patent application WO 2020074915 for the iCaspase9 transgenic chicken. The University of Edinburgh is the applicant. The remaining authors declare no competing interests.
Figures




References
-
- Ritchie, H. & Roser, M. https://ourworldindata.org/meat-production#number-of-animals-slaughtered (accessed 2020).
-
- Melesse A. Significance of scavenging chicken production in the rural community of Africa for enhanced food security. World Poult. Sci. J. 2014;70:593–606. doi: 10.1017/S0043933914000646. - DOI
-
- Alders RG, Pym RAE. Village poultry: still important to millions, eight thousand years after domestication. World Poult. Sci. J. 2009;65:181–190. doi: 10.1017/S0043933909000117. - DOI
-
- Dad-IS.
Publication types
MeSH terms
Grants and funding
- BBS/E/D/20320000/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011895/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P013759/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P013732/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

