High-throughput measurement of fibroblast rhythms reveals genetic heritability of circadian phenotypes in diversity outbred mice and their founder strains
- PMID: 33510298
- PMCID: PMC7843998
- DOI: 10.1038/s41598-021-82069-8
High-throughput measurement of fibroblast rhythms reveals genetic heritability of circadian phenotypes in diversity outbred mice and their founder strains
Abstract
Circadian variability is driven by genetics and Diversity Outbred (DO) mice is a powerful tool for examining the genetics of complex traits because their high genetic and phenotypic diversity compared to conventional mouse crosses. The DO population combines the genetic diversity of eight founder strains including five common inbred and three wild-derived strains. In DO mice and their founders, we established a high-throughput system to measure cellular rhythms using in vitro preparations of skin fibroblasts. Among the founders, we observed strong heritability for rhythm period, robustness, phase and amplitude. We also found significant sex and strain differences for these rhythms. Extreme differences in period for molecular and behavioral rhythms were found between the inbred A/J strain and the wild-derived CAST/EiJ strain, where A/J had the longest period and CAST/EiJ had the shortest. In addition, we measured cellular rhythms in 329 DO mice, which displayed far greater phenotypic variability than the founders-80% of founders compared to only 25% of DO mice had periods of ~ 24 h. Collectively, our findings demonstrate that genetic diversity contributes to phenotypic variability in circadian rhythms, and high-throughput characterization of fibroblast rhythms in DO mice is a tractable system for examining the genetics of circadian traits.
Conflict of interest statement
The authors declare no competing interests.
Figures
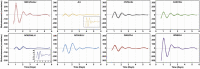
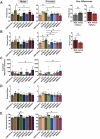
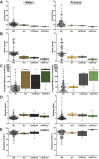

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

