Chemical systems biology reveals mechanisms of glucocorticoid receptor signaling
- PMID: 33510451
- PMCID: PMC8783757
- DOI: 10.1038/s41589-020-00719-w
Chemical systems biology reveals mechanisms of glucocorticoid receptor signaling
Abstract
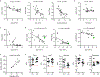
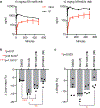
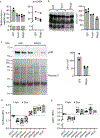
Glucocorticoids display remarkable anti-inflammatory activity, but their use is limited by on-target adverse effects including insulin resistance and skeletal muscle atrophy. We used a chemical systems biology approach, ligand class analysis, to examine ligands designed to modulate glucocorticoid receptor activity through distinct structural mechanisms. These ligands displayed diverse activity profiles, providing the variance required to identify target genes and coregulator interactions that were highly predictive of their effects on myocyte glucose disposal and protein balance. Their anti-inflammatory effects were linked to glucose disposal but not muscle atrophy. This approach also predicted selective modulation in vivo, identifying compounds that were muscle-sparing or anabolic for protein balance and mitochondrial potential. Ligand class analysis defined the mechanistic links between the ligand-receptor interface and ligand-driven physiological outcomes, a general approach that can be applied to any ligand-regulated allosteric signaling system.
Conflict of interest statement
Figures










References
Methods-only References
-
- Heikkinen S, Argmann CA, Champy MF & Auwerx J Evaluation of glucose homeostasis. Curr Protoc Mol Biol Chapter 29, Unit 29B 3 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

