Whole Genome Analysis of African G12P[6] and G12P[8] Rotaviruses Provides Evidence of Porcine-Human Reassortment at NSP2, NSP3, and NSP4
- PMID: 33510725
- PMCID: PMC7835662
- DOI: 10.3389/fmicb.2020.604444
Whole Genome Analysis of African G12P[6] and G12P[8] Rotaviruses Provides Evidence of Porcine-Human Reassortment at NSP2, NSP3, and NSP4
Abstract
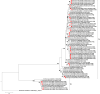
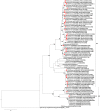
Group A rotaviruses (RVA) represent the most common cause of pediatric gastroenteritis in children <5 years, worldwide. There has been an increase in global detection and reported cases of acute gastroenteritis caused by RVA genotype G12 strains, particularly in Africa. This study sought to characterize the genomic relationship between African G12 strains and determine the possible origin of these strains. Whole genome sequencing of 34 RVA G12P[6] and G12P[8] strains detected from the continent including southern (South Africa, Zambia, Zimbabwe), eastern (Ethiopia, Uganda), central (Cameroon), and western (Togo) African regions, were sequenced using the Ion Torrent PGM method. The majority of the strains possessed a Wa-like backbone with consensus genotype constellation of G12-P[6]/P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1, while a single strain from Ethiopia displayed a DS-1-like genetic constellation of G12-P[6]-I2-R2-C2-M2-A2-N2-T2-E2-H2. In addition, three Ethiopian and one South African strains exhibited a genotype 2 reassortment of the NSP3 gene, with genetic constellation of G12-P[8]-I1-R1-C1-M1-A1-N1-T2-E1-H1. Overall, 10 gene segments (VP1-VP4, VP6, and NSP1-NSP5) of African G12 strains were determined to be genetically related to cognate gene sequences from globally circulating human Wa-like G12, G9, and G1 strains with nucleotide (amino acid) identities in the range of 94.1-99.9% (96.5-100%), 88.5-98.5% (93-99.1%), and 89.8-99.0% (88.7-100%), respectively. Phylogenetic analysis showed that the Ethiopian G12P[6] possessing a DS-1-like backbone consistently clustered with G2P[4] strains from Senegal and G3P[6] from Ethiopia with the VP1, VP2, VP6, and NSP1-NSP4 genes. Notably, the NSP2, NSP3, and NSP4 of most of the study strains exhibited the closest relationship with porcine strains suggesting the occurrence of reassortment between human and porcine strains. Our results add to the understanding of potential roles that interspecies transmission play in generating human rotavirus diversity through reassortment events and provide insights into the evolutionary dynamics of G12 strains spreading across selected sub-Saharan Africa regions.
Keywords: Africa; genotype 12; group A rotaviruses; next generation sequencing; reassortment; whole genome sequencing.
Copyright © 2021 Mokoena, Esona, Seheri, Nyaga, Magagula, Mukaratirwa, Mulindwa, Abebe, Boula, Tsolenyanu, Simwaka, Rakau, Peenze, Mwenda, Mphahlele and Steele.
Conflict of interest statement
AS was employed by the Bill & Melinda Gates Foundation which is working on rotavirus vaccine development and deployment. Understanding rotavirus strain diversity and evolution, including under possible vaccine pressure, is an important goal. The Bill & Melinda Gates Foundation partially funded the African Enteric Viruses Genome Initiative (AEVGI) at the University of Free State, but was not involved in sample selection or the laboratory methodology used. AS was engaged in the interpretation of the data and the drafting and approval of the manuscript as the senior corresponding author. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abebe A., Getahun M., Mapaseka S. L., Beyene B., Assefa E., Teshome B., et al. (2018). Impact of rotavirus vaccine introduction and genotypic characteristics of rotavirus strains in children less than 5years of age with gastroenteritis in Ethiopia: 2011-2016. Vaccine 36 7043–7047. 10.1016/j.vaccine.2018.09.048 - DOI - PubMed
-
- Bányai K., László B., Duque J., Steele A. D., Nelson E. A., Gentsch J. R., et al. (2012). Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 30 A122–A130. 10.1016/j.vaccine.2011.09.111 - DOI - PubMed
-
- Bwogi J., Jere K. C., Karamagi C., Byarugaba D. K., Namuwulya P., Baliraine F. N., et al. (2017). Whole genome analysis of selected human and animal rotaviruses identified in Uganda from 2012 to 2014 reveals complex genome reassortment events between human, bovine, caprine and porcine strains. PLoS One 12:e0178855. 10.1371/journal.pone.0178855 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

