Barley Anther and Meiocyte Transcriptome Dynamics in Meiotic Prophase I
- PMID: 33510760
- PMCID: PMC7835676
- DOI: 10.3389/fpls.2020.619404
Barley Anther and Meiocyte Transcriptome Dynamics in Meiotic Prophase I
Abstract
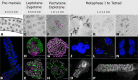
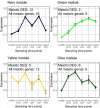
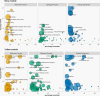
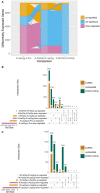
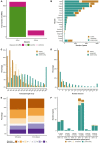
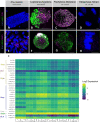
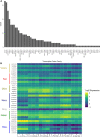
In flowering plants, successful germinal cell development and meiotic recombination depend upon a combination of environmental and genetic factors. To gain insights into this specialized reproductive development program we used short- and long-read RNA-sequencing (RNA-seq) to study the temporal dynamics of transcript abundance in immuno-cytologically staged barley (Hordeum vulgare) anthers and meiocytes. We show that the most significant transcriptional changes in anthers occur at the transition from pre-meiosis to leptotene-zygotene, which is followed by increasingly stable transcript abundance throughout prophase I into metaphase I-tetrad. Our analysis reveals that the pre-meiotic anthers are enriched in long non-coding RNAs (lncRNAs) and that entry to meiosis is characterized by their robust and significant down regulation. Intriguingly, only 24% of a collection of putative meiotic gene orthologs showed differential transcript abundance in at least one stage or tissue comparison. Argonautes, E3 ubiquitin ligases, and lys48 specific de-ubiquitinating enzymes were enriched in prophase I meiocyte samples. These developmental, time-resolved transcriptomes demonstrate remarkable stability in transcript abundance in meiocytes throughout prophase I after the initial and substantial reprogramming at meiosis entry and the complexity of the regulatory networks involved in early meiotic processes.
Keywords: anther; argonaute; barley; lncRNAs; meiocyte; meiosis; transcriptome; ubiquitin.
Copyright © 2021 Barakate, Orr, Schreiber, Colas, Lewandowska, McCallum, Macaulay, Morris, Arrieta, Hedley, Ramsay and Waugh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

