Transcriptome annotation in the cloud: complexity, best practices, and cost
- PMID: 33511996
- PMCID: PMC7845158
- DOI: 10.1093/gigascience/giaa163
Transcriptome annotation in the cloud: complexity, best practices, and cost
Abstract
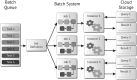
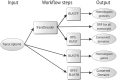
Background: The NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) initiative provides NIH-funded researchers cost-effective access to commercial cloud providers, such as Amazon Web Services (AWS) and Google Cloud Platform (GCP). These cloud providers represent an alternative for the execution of large computational biology experiments like transcriptome annotation, which is a complex analytical process that requires the interrogation of multiple biological databases with several advanced computational tools. The core components of annotation pipelines published since 2012 are BLAST sequence alignments using annotated databases of both nucleotide or protein sequences almost exclusively with networked on-premises compute systems.
Findings: We compare multiple BLAST sequence alignments using AWS and GCP. We prepared several Jupyter Notebooks with all the code required to submit computing jobs to the batch system on each cloud provider. We consider the consequence of the number of query transcripts in input files and the effect on cost and processing time. We tested compute instances with 16, 32, and 64 vCPUs on each cloud provider. Four classes of timing results were collected: the total run time, the time for transferring the BLAST databases to the instance local solid-state disk drive, the time to execute the CWL script, and the time for the creation, set-up, and release of an instance. This study aims to establish an estimate of the cost and compute time needed for the execution of multiple BLAST runs in a cloud environment.
Conclusions: We demonstrate that public cloud providers are a practical alternative for the execution of advanced computational biology experiments at low cost. Using our cloud recipes, the BLAST alignments required to annotate a transcriptome with ∼500,000 transcripts can be processed in <2 hours with a compute cost of ∼$200-$250. In our opinion, for BLAST-based workflows, the choice of cloud platform is not dependent on the workflow but, rather, on the specific details and requirements of the cloud provider. These choices include the accessibility for institutional use, the technical knowledge required for effective use of the platform services, and the availability of open source frameworks such as APIs to deploy the workflow.
Published by Oxford University Press on behalf of GigaScience 2021.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- NIH STRIDES Initiative. https://cloud.cit.nih.gov/.Accessed 19 January, 2021
-
- SRA in the Cloud. https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud/. Accessed 19 January, 2021
-
- Official NCBI BLAST+ Docker Image Documentation. https://github.com/ncbi/blast_plus_docs. Accessed 19 January, 2021
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

