Temporally integrated single cell RNA sequencing analysis of PBMC from experimental and natural primary human DENV-1 infections
- PMID: 33513191
- PMCID: PMC7875406
- DOI: 10.1371/journal.ppat.1009240
Temporally integrated single cell RNA sequencing analysis of PBMC from experimental and natural primary human DENV-1 infections
Abstract
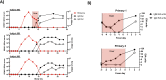
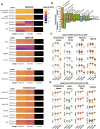
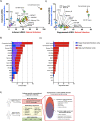
Dengue human infection studies present an opportunity to address many longstanding questions in the field of flavivirus biology. However, limited data are available on how the immunological and transcriptional response elicited by an attenuated challenge virus compares to that associated with a wild-type DENV infection. To determine the kinetic transcriptional signature associated with experimental primary DENV-1 infection and to assess how closely this profile correlates with the transcriptional signature accompanying natural primary DENV-1 infection, we utilized scRNAseq to analyze PBMC from individuals enrolled in a DENV-1 human challenge study and from individuals experiencing a natural primary DENV-1 infection. While both experimental and natural primary DENV-1 infection resulted in overlapping patterns of inflammatory gene upregulation, natural primary DENV-1 infection was accompanied with a more pronounced suppression in gene products associated with protein translation and mitochondrial function, principally in monocytes. This suggests that the immune response elicited by experimental and natural primary DENV infection are similar, but that natural primary DENV-1 infection has a more pronounced impact on basic cellular processes to induce a multi-layered anti-viral state.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests:A.T.W reports grants from Military Infectious Disease Research Program, during the conduct of the study. A.L.R. reports grants from National Institute of Allergy and Infectious Diseases, during the conduct of the study. S.J.T reports other support from US DoD, other support from GSK, during the conduct of the study; personal fees and other support from GSK Vaccines, personal fees and other support from Takeda, personal fees and other support from Merck, personal fees and other support from PrimeVax, personal fees and other support from Themisbio, personal fees and other support from Chugai Pharma, personal fees and other support from Cormac Life Sciences, personal fees and other support from HHS NVPO / Tunnel Govt Services, personal fees and other support from Janssen, other support from GreenMark Partners. In addition, S.J.T has a patent US10086061B2 (combined flavivirus vaccines) issued. J.R.C reports grants from the Congressionally Directed Medical Research Program during the conduct of the study. All other authors have nothing to disclose.
Figures


References
-
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, et al. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol 1984;120(5):653–69. Epub 1984/11/01. 10.1093/oxfordjournals.aje.a113932 . - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

