Single-cell transcriptomics reveal the intratumoral landscape of infiltrated T-cell subpopulations in oral squamous cell carcinoma
- PMID: 33513276
- PMCID: PMC8024729
- DOI: 10.1002/1878-0261.12910
Single-cell transcriptomics reveal the intratumoral landscape of infiltrated T-cell subpopulations in oral squamous cell carcinoma
Abstract
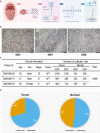
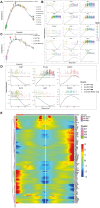
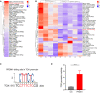
Systematic analysis of tumor-infiltrating lymphocytes is essential for the development of new cancer treatments and the prediction of clinical responses to immunotherapy. Immunomodulatory drugs are used for the treatment of oral squamous cell carcinoma (OSCC), depending on immune infiltration profiles of the tumor microenvironment. In this study, we isolated 11,866 single T cells from tumors and paired adjacent normal tissues of three patients with OSCC. Using single-cell RNA sequencing, we identified 14 distinct T-cell subpopulations within the tumors and 5 T-cell subpopulations in the adjacent normal tissues and delineated their developmental trajectories. Exhausted CD8+ T cells and regulatory CD4+ T cells (CD4+ Tregs) were enriched in OSCC tumors, potentially linked to tumor immunosuppression. Programmed death protein 1 (PD-1) and cytotoxic T lymphocyte-associated protein 4 (CTLA4) were identified as marker genes in exhausted CD8+ T cells, whereas forkhead box P3 (FOXP3) and CTLA4 were identified as markers of CD4+ Tregs. Furthermore, our data revealed that thymocyte selection-associated high-mobility group box (TOX) may be a key regulator of T-cell dysfunction in the OSCC microenvironment. Overexpression of TOX upregulated expression of genes related to T-cell dysfunction. In vitro experiments demonstrated that cytotoxic activity and proliferation efficiency of CD8+ T cells overexpressing PD-1 or TOX were reduced. Notable, the transcription factor PRDM1 was found to transactivate TOX expression via a binding motif in the TOX promoter. Our findings provide valuable insight into the functional states and heterogeneity of T-cell populations in OSCC that could advance the development of novel therapeutic strategies.
Keywords: T-cell exhaustion; cancer immunology; oral squamous cell carcinoma; single-cell sequencing; tumor-infiltrating lymphocytes.
© 2021 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Weinberg RA (2014) Coming full circle—from endless complexity to simplicity and back again. Cell 157, 267–271. - PubMed
-
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin Dm, Piñeros M, Znaor A & Bray F (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144, 1941–1953. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

