Gene Therapy of Dominant CRX-Leber Congenital Amaurosis using Patient Stem Cell-Derived Retinal Organoids
- PMID: 33513359
- PMCID: PMC7878833
- DOI: 10.1016/j.stemcr.2020.12.018
Gene Therapy of Dominant CRX-Leber Congenital Amaurosis using Patient Stem Cell-Derived Retinal Organoids
Abstract
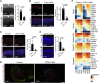
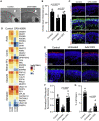
Mutations in the photoreceptor transcription factor gene cone-rod homeobox (CRX) lead to distinct retinopathy phenotypes, including early-onset vision impairment in dominant Leber congenital amaurosis (LCA). Using induced pluripotent stem cells (iPSCs) from a patient with CRX-I138fs48 mutation, we established an in vitro model of CRX-LCA in retinal organoids that showed defective photoreceptor maturation by histology and gene profiling, with diminished expression of visual opsins. Adeno-associated virus (AAV)-mediated CRX gene augmentation therapy partially restored photoreceptor phenotype and expression of phototransduction-related genes as determined by single-cell RNA-sequencing. Retinal organoids derived from iPSCs of a second dominant CRX-LCA patient carrying K88N mutation revealed the loss of opsin expression as a common phenotype, which was alleviated by AAV-mediated augmentation of CRX. Our studies provide a proof-of-concept for developing gene therapy of dominant CRX-LCA and other CRX retinopathies.
Keywords: 3-D organoids; AAV; disease modeling; iPSC; pluripotent stem cells; retinal degeneration; scRNA-seq; therapy; transcription factor; transcriptome.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Ahmed C.M., Dwyer B.T., Romashko A., Van Adestine S., Park E.H., Lou Z., Welty D., Josiah S., Savinainen A., Zhang B. SRD005825 acts as a pharmacologic chaperone of opsin and promotes survival of photoreceptors in an animal model of autosomal dominant retinitis pigmentosa. Transl. Vis. Sci. Technol. 2019;8:30. - PMC - PubMed
-
- Apte R.S. Gene therapy for retinal degeneration. Cell. 2018;173:5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

