The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies
- PMID: 33514027
- PMCID: PMC7911644
- DOI: 10.3390/genes12020184
The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies
Abstract
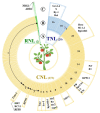
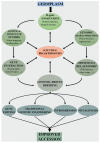
Tomato (Solanum lycopersicum L.) is a model system for studying the molecular basis of resistance in plants. The investigation of evolutionary dynamics of tomato resistance (R)-loci provides unique opportunities for identifying factors that promote or constrain genome evolution. Nucleotide-binding domain and leucine-rich repeat (NB-LRR) receptors belong to one of the most plastic and diversified families. The vast amount of genomic data available for Solanaceae and wild tomato relatives provides unprecedented insights into the patterns and mechanisms of evolution of NB-LRR genes. Comparative analysis remarked a reshuffling of R-islands on chromosomes and a high degree of adaptive diversification in key R-loci induced by species-specific pathogen pressure. Unveiling NB-LRR natural variation in tomato and in other Solanaceae species offers the opportunity to effectively exploit genetic diversity in genomic-driven breeding programs with the aim of identifying and introducing new resistances in tomato cultivars. Within this motivating context, we reviewed the repertoire of NB-LRR genes available for tomato improvement with a special focus on signatures of adaptive processes. This issue is still relevant and not thoroughly investigated. We believe that the discovery of mechanisms involved in the generation of a gene with new resistance functions will bring great benefits to future breeding strategies.
Keywords: NLR genes; R-genes; evolutionary dynamics; gene clusters; genomic-driven breeding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Peralta I.E., Spooner D.M., Knapp S. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae) Syst. Bot. Monogr. 2008;84:1–186.
-
- Arie T., Takahashi H., Kodama M., Teraoka T. Tomato as a model plant for plant-pathogen interactions. Plant Biotechnol. 2007;24:135–147. doi: 10.5511/plantbiotechnology.24.135. - DOI
-
- Andolfo G., Sanseverino W., Rombauts S., Van de Peer Y., Bradeen J.M., Carputo D., Frusciante L., Ercolano M.R. Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol. 2013;197:223–237. doi: 10.1111/j.1469-8137.2012.04380.x. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

