Genomics Insights into Pseudomonas sp. CG01: An Antarctic Cadmium-Resistant Strain Capable of Biosynthesizing CdS Nanoparticles Using Methionine as S-Source
- PMID: 33514061
- PMCID: PMC7912247
- DOI: 10.3390/genes12020187
Genomics Insights into Pseudomonas sp. CG01: An Antarctic Cadmium-Resistant Strain Capable of Biosynthesizing CdS Nanoparticles Using Methionine as S-Source
Abstract
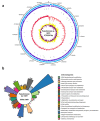
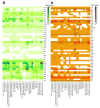
Here, we present the draft genome sequence of Pseudomonas sp. GC01, a cadmium-resistant Antarctic bacterium capable of biosynthesizing CdS fluorescent nanoparticles (quantum dots, QDs) employing a unique mechanism involving the production of methanethiol (MeSH) from methionine (Met). To explore the molecular/metabolic components involved in QDs biosynthesis, we conducted a comparative genomic analysis, searching for the genes related to cadmium resistance and sulfur metabolic pathways. The genome of Pseudomonas sp. GC01 has a 4,706,645 bp size with a 58.61% G+C content. Pseudomonas sp. GC01 possesses five genes related to cadmium transport/resistance, with three P-type ATPases (cadA, zntA, and pbrA) involved in Cd-secretion that could contribute to the extracellular biosynthesis of CdS QDs. Furthermore, it exhibits genes involved in sulfate assimilation, cysteine/methionine synthesis, and volatile sulfur compounds catabolic pathways. Regarding MeSH production from Met, Pseudomonas sp. GC01 lacks the genes E4.4.1.11 and megL for MeSH generation. Interestingly, despite the absence of these genes, Pseudomonas sp. GC01 produces high levels of MeSH. This is probably associated with the metC gene that also produces MeSH from Met in bacteria. This work is the first report of the potential genes involved in Cd resistance, sulfur metabolism, and the process of MeSH-dependent CdS QDs bioproduction in Pseudomonas spp. strains.
Keywords: Antarctic bacteria; comparative genomics; nanoparticle biosynthesis; volatile sulfur compounds.
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Wasley J., Robinson S.A., Lovelock C.E., Popp M. Climate change manipulations show Antarctic flora is more strongly affected by elevated nutrients than water. Glob. Chang. Biol. 2006;12:1800–1812. doi: 10.1111/j.1365-2486.2006.01209.x. - DOI
-
- Nichols D.S., Sanderson K., Buia A., Van de Kamp J., Holloway P., Bowman J.P., Smith M., Nichols C.M., Nichols P.D., McMeekin T.A. Bioprospecting and biotechnology in Antarctica. In: Jabour-Green J., Haward M., editors. The Antarctic: Past, Present and Future, Antarctic CRC Research Report 28. Antarctic CRC; Hobart, Australia: 2002. pp. 85–103.
-
- Romoli R., Papaleo M., de Pascale D., Tutino M., Michaud L., LoGiudice A., Bartolucci G. Characterization of the volatile profile of Antarctic bacteria by using solid-phase microextraction– gas chromatography-mass spectrometry. J. Mass Spectrom. 2011;46:1051–1059. doi: 10.1002/jms.1987. - DOI - PubMed
-
- Papaleo M., Fondi M., Maida I., Perrin E., Lo Giudice A., Michaud L., Mangano S., Bartolucci G., Romoli R., Fani R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012;30:272–293. doi: 10.1016/j.biotechadv.2011.06.011. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

