The skeletome of the red coral Corallium rubrum indicates an independent evolution of biomineralization process in octocorals
- PMID: 33514311
- PMCID: PMC7853314
- DOI: 10.1186/s12862-020-01734-0
The skeletome of the red coral Corallium rubrum indicates an independent evolution of biomineralization process in octocorals
Abstract
Background: The process of calcium carbonate biomineralization has arisen multiple times during metazoan evolution. In the phylum Cnidaria, biomineralization has mostly been studied in the subclass Hexacorallia (i.e. stony corals) in comparison to the subclass Octocorallia (i.e. red corals); the two diverged approximately 600 million years ago. The precious Mediterranean red coral, Corallium rubrum, is an octocorallian species, which produces two distinct high-magnesium calcite biominerals, the axial skeleton and the sclerites. In order to gain insight into the red coral biomineralization process and cnidarian biomineralization evolution, we studied the protein repertoire forming the organic matrix (OM) of its two biominerals.
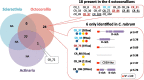
Results: We combined High-Resolution Mass Spectrometry and transcriptome analysis to study the OM composition of the axial skeleton and the sclerites. We identified a total of 102 OM proteins, 52 are found in the two red coral biominerals with scleritin being the most abundant protein in each fraction. Contrary to reef building corals, the red coral organic matrix possesses a large number of collagen-like proteins. Agrin-like glycoproteins and proteins with sugar-binding domains are also predominant. Twenty-seven and 23 proteins were uniquely assigned to the axial skeleton and the sclerites, respectively. The inferred regulatory function of these OM proteins suggests that the difference between the two biominerals is due to the modeling of the matrix network, rather than the presence of specific structural components. At least one OM component could have been horizontally transferred from prokaryotes early during Octocorallia evolution.
Conclusion: Our results suggest that calcification of the red coral axial skeleton likely represents a secondary calcification of an ancestral gorgonian horny axis. In addition, the comparison with stony coral skeletomes highlighted the low proportion of similar proteins between the biomineral OMs of hexacorallian and octocorallian corals, suggesting an independent acquisition of calcification in anthozoans.
Keywords: Axial skeleton; Biomineralization; Corallium rubrum; Evolution; Organic matrix; Proteomics; Sclerites.
Conflict of interest statement
The authors declare that there have no competing interests.
Figures

Similar articles
-
New Non-Bilaterian Transcriptomes Provide Novel Insights into the Evolution of Coral Skeletomes.Genome Biol Evol. 2019 Nov 1;11(11):3068-3081. doi: 10.1093/gbe/evz199. Genome Biol Evol. 2019. PMID: 31518412 Free PMC article.
-
Comparative Proteomics of Octocoral and Scleractinian Skeletomes and the Evolution of Coral Calcification.Genome Biol Evol. 2020 Sep 1;12(9):1623-1635. doi: 10.1093/gbe/evaa162. Genome Biol Evol. 2020. PMID: 32761183 Free PMC article.
-
Characterization of the proteinaceous skeletal organic matrix from the precious coral Corallium konojoi.Proteomics. 2014 Nov;14(21-22):2600-6. doi: 10.1002/pmic.201300519. Epub 2014 Oct 3. Proteomics. 2014. PMID: 25044550
-
How corals made rocks through the ages.Glob Chang Biol. 2020 Jan;26(1):31-53. doi: 10.1111/gcb.14912. Epub 2019 Dec 14. Glob Chang Biol. 2020. PMID: 31696576 Free PMC article. Review.
-
The Skeleton and Biomineralization Mechanism as Part of the Innate Immune System of Stony Corals.Front Immunol. 2022 Feb 25;13:850338. doi: 10.3389/fimmu.2022.850338. eCollection 2022. Front Immunol. 2022. PMID: 35281045 Free PMC article. Review.
Cited by
-
An alternative and effective method for extracting skeletal organic matrix adapted to the red coral Corallium rubrum.Biol Open. 2022 Oct 15;11(10):bio059536. doi: 10.1242/bio.059536. Epub 2022 Oct 14. Biol Open. 2022. PMID: 36178163 Free PMC article.
-
Chromosome-level genome assembly of the black widow spider Latrodectus elegans illuminates composition and evolution of venom and silk proteins.Gigascience. 2022 May 25;11:giac049. doi: 10.1093/gigascience/giac049. Gigascience. 2022. PMID: 35639632 Free PMC article.
-
Diversity and cross-species transmission of viruses in a remote island ecosystem: implications for wildlife conservation.Virus Evol. 2024 Dec 14;11(1):veae113. doi: 10.1093/ve/veae113. eCollection 2025. Virus Evol. 2024. PMID: 39802822 Free PMC article.
-
Revealing Elasmobranch Distributions in Turbid Coastal Waters: Insights From Environmental DNA and Particle Tracking.Ecol Evol. 2025 Jan 24;15(1):e70857. doi: 10.1002/ece3.70857. eCollection 2025 Jan. Ecol Evol. 2025. PMID: 39867497 Free PMC article.
-
Hippo-vgll3 signaling may contribute to sex differences in Atlantic salmon maturation age via contrasting adipose dynamics.Biol Sex Differ. 2025 Apr 2;16(1):23. doi: 10.1186/s13293-025-00705-8. Biol Sex Differ. 2025. PMID: 40176157 Free PMC article.
References
-
- Williams RJP. An introduction to biominerals and the role of organic molecules in their formation. Phil Trans R Soc Lond B. 1984;304:411–424. doi: 10.1098/rstb.1984.0035. - DOI
-
- Watabe N, Kingsley RJ. Calcification in octocorals. In: Suga S, Watabe N, editors. Hard tissue mineralization and demineralization. Tokyo: Springer; 1992. pp. 127–147.
-
- Kingsley RJ, Watabe N. Ultrastructure of the axial region in Leptogorgia virgulata (Cnidaria: Gorgonaceae) Trans Am Microsc Soc. 1982;101:325. doi: 10.2307/3225750. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
