SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells
- PMID: 33514628
- PMCID: PMC8103705
- DOI: 10.1128/JVI.02415-20
SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells
Abstract
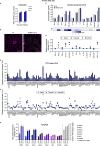
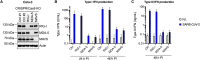
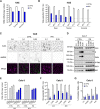
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of coronavirus disease 19 (COVID-19), which ranges from mild respiratory symptoms to acute respiratory distress syndrome, and death in the most severe cases. Immune dysregulation with altered innate cytokine responses is thought to contribute to disease severity. Here, we characterized in depth host cell responses against SARS-CoV-2 in primary human airway epithelia (HAE) and immortalized cell lines. Our results demonstrate that primary HAE and model cells elicit a robust induction of type I and III interferons (IFNs). Importantly, we show for the first time that melanoma differentiation associated gene (MDA)-5 is the main sensor of SARS-CoV-2 in lung cells. IFN exposure strongly inhibited viral replication and de novo production of infectious virions. However, despite high levels of IFNs produced in response to SARS-CoV-2 infection, the IFN response was unable to control viral replication in lung cells, contrary to what was previously reported in intestinal epithelial cells. Altogether, these results highlight the complex and ambiguous interplay between viral replication and the timing of IFN responses.IMPORTANCE Mammalian cells express sensors able to detect specific features of pathogens and induce the interferon response, which is one of the first line of defenses against viruses and help controlling viral replication. The mechanisms and impact of SARS-CoV-2 sensing in lung epithelial cells remained to be deciphered. In this study, we report that despite a high production of type I and III interferons specifically induced by MDA-5-mediated sensing of SARS-CoV-2, primary and immortalized lung epithelial cells are unable to control viral replication. However, exogenous interferons potently inhibited replication, if provided early upon viral exposure. A better understanding of the ambiguous interplay between the interferon response and SARS-CoV-2 replication is essential to guide future therapeutical interventions.
Copyright © 2021 American Society for Microbiology.
Figures






References
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Zhong N, Zheng B, Li Y, Poon L, Xie Z, Chan K, Li P, Tan S, Chang Q, Xie J, Liu X, Xu J, Li D, Yuen K, Peiris J, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. - DOI - PMC - PubMed
-
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM, Berger A, Burguière A-M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J-C, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk H-D, Osterhaus ADME, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

