Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare
- PMID: 33514699
- PMCID: PMC7846756
- DOI: 10.1038/s41467-021-20910-4
Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare
Abstract
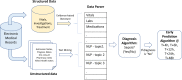
Sepsis is a leading cause of death in hospitals. Early prediction and diagnosis of sepsis, which is critical in reducing mortality, is challenging as many of its signs and symptoms are similar to other less critical conditions. We develop an artificial intelligence algorithm, SERA algorithm, which uses both structured data and unstructured clinical notes to predict and diagnose sepsis. We test this algorithm with independent, clinical notes and achieve high predictive accuracy 12 hours before the onset of sepsis (AUC 0.94, sensitivity 0.87 and specificity 0.87). We compare the SERA algorithm against physician predictions and show the algorithm's potential to increase the early detection of sepsis by up to 32% and reduce false positives by up to 17%. Mining unstructured clinical notes is shown to improve the algorithm's accuracy compared to using only clinical measures for early warning 12 to 48 hours before the onset of sepsis.
Conflict of interest statement
The authors declare no competing interests.
Figures

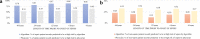


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

