A novel CD4+ CTL subtype characterized by chemotaxis and inflammation is involved in the pathogenesis of Graves' orbitopathy
- PMID: 33514849
- PMCID: PMC8027210
- DOI: 10.1038/s41423-020-00615-2
A novel CD4+ CTL subtype characterized by chemotaxis and inflammation is involved in the pathogenesis of Graves' orbitopathy
Abstract
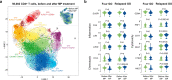
Graves' orbitopathy (GO), the most severe manifestation of Graves' hyperthyroidism (GH), is an autoimmune-mediated inflammatory disorder, and treatments often exhibit a low efficacy. CD4+ T cells have been reported to play vital roles in GO progression. To explore the pathogenic CD4+ T cell types that drive GO progression, we applied single-cell RNA sequencing (scRNA-Seq), T cell receptor sequencing (TCR-Seq), flow cytometry, immunofluorescence and mixed lymphocyte reaction (MLR) assays to evaluate CD4+ T cells from GO and GH patients. scRNA-Seq revealed the novel GO-specific cell type CD4+ cytotoxic T lymphocytes (CTLs), which are characterized by chemotactic and inflammatory features. The clonal expansion of this CD4+ CTL population, as demonstrated by TCR-Seq, along with their strong cytotoxic response to autoantigens, localization in orbital sites, and potential relationship with disease relapse provide strong evidence for the pathogenic roles of GZMB and IFN-γ-secreting CD4+ CTLs in GO. Therefore, cytotoxic pathways may become potential therapeutic targets for GO.
Keywords: CD4+ cytotoxic T lymphocytes; Graves’ orbitopathy; single-cell RNA sequencing.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Smith TJ, Hegedüs L. Graves’ Disease. N. Engl. J. Med. 2016;375:1552–1565. - PubMed
-
- Taylor PN, et al. New insights into the pathogenesis and nonsurgical management of Graves orbitopathy. Nat. Rev. Endocrinol. 2020;16:104–116. - PubMed
-
- Ye X, et al. Efficacy and safety of mycophenolate mofetil in patients with active moderate-to-severe Graves’ orbitopathy. Clin. Endocrinol. 2017;86:247–255. - PubMed
-
- Kahaly GJ, et al. Mycophenolate plus methylprednisolone versus methylprednisolone alone in active, moderate-to-severe Graves’ orbitopathy (MINGO): a randomised, observer-masked, multicentre trial. Lancet Diabetes Endocrinol. 2018;6:287–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

