Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage
- PMID: 33514850
- PMCID: PMC7846785
- DOI: 10.1038/s41598-021-82164-w
Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage
Abstract
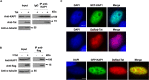
HIV-1 latency generates reservoirs that prevent viral eradication by the current therapies. To find strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent repressor of gene transcription. Here we report that KAP1 represses HIV-1 gene expression in myeloid cells including microglial cells, the major reservoir of the central nervous system. Mechanistically, KAP1 interacts and colocalizes with the viral transactivator Tat to promote its degradation via the proteasome pathway and repress HIV-1 gene expression. In myeloid models of latent HIV-1 infection, the depletion of KAP1 increased viral gene elongation and reactivated HIV-1 expression. Bound to the latent HIV-1 promoter, KAP1 associates and cooperates with CTIP2, a key epigenetic silencer of HIV-1 expression in microglial cells. In addition, Tat and CTIP2 compete for KAP1 binding suggesting a dynamic modulation of the KAP1 cellular partners upon HIV-1 infection. Altogether, our results suggest that KAP1 contributes to the establishment and the persistence of HIV-1 latency in myeloid cells.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

