Colonization and genetic diversification processes of Leishmania infantum in the Americas
- PMID: 33514858
- PMCID: PMC7846609
- DOI: 10.1038/s42003-021-01658-5
Colonization and genetic diversification processes of Leishmania infantum in the Americas
Abstract
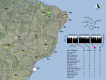
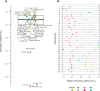
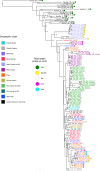
Leishmania infantum causes visceral leishmaniasis, a deadly vector-borne disease introduced to the Americas during the colonial era. This non-native trypanosomatid parasite has since established widespread transmission cycles using alternative vectors, and human infection has become a significant concern to public health, especially in Brazil. A multi-kilobase deletion was recently detected in Brazilian L. infantum genomes and is suggested to reduce susceptibility to the anti-leishmanial drug miltefosine. We show that deletion-carrying strains occur in at least 15 Brazilian states and describe diversity patterns suggesting that these derive from common ancestral mutants rather than from recurrent independent mutation events. We also show that the deleted locus and associated enzymatic activity is restored by hybridization with non-deletion type strains. Genetic exchange appears common in areas of secondary contact but also among closely related parasites. We examine demographic and ecological scenarios underlying this complex L. infantum population structure and discuss implications for disease control.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

