Revealing nascent RNA processing dynamics with nano-COP
- PMID: 33514943
- PMCID: PMC8713461
- DOI: 10.1038/s41596-020-00469-y
Revealing nascent RNA processing dynamics with nano-COP
Abstract
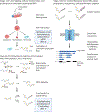
During maturation, eukaryotic precursor RNAs undergo processing events including intron splicing, 3'-end cleavage, and polyadenylation. Here we describe nanopore analysis of co-transcriptional processing (nano-COP), a method for probing the timing and patterns of RNA processing. An extension of native elongating transcript sequencing, which quantifies transcription genome-wide through short-read sequencing of nascent RNA 3' ends, nano-COP uses long-read nascent RNA sequencing to observe global patterns of RNA processing. First, nascent RNA is stringently purified through a combination of 4-thiouridine metabolic labeling and cellular fractionation. In contrast to cDNA or short-read-based approaches relying on reverse transcription or amplification, the sample is sequenced directly through nanopores to reveal the native context of nascent RNA. nano-COP identifies both active transcription sites and splice isoforms of single RNA molecules during synthesis, providing insight into patterns of intron removal and the physical coupling between transcription and splicing. The nano-COP protocol yields data within 3 d.
Figures

References
-
- Mariman EC, van Beek-Reinders RJ & van Venrooij WJ Alternative splicing pathways exist in the formation of adenoviral late messenger RNAs. J. Mol. Biol 163, 239–256 (1983). - PubMed
-
- Beyer AL & Osheim YN Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2, 754–765 (1988). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

