Detailed evaluation of pyruvate dehydrogenase complex inhibition in simulated exercise conditions
- PMID: 33515599
- PMCID: PMC8008327
- DOI: 10.1016/j.bpj.2021.01.018
Detailed evaluation of pyruvate dehydrogenase complex inhibition in simulated exercise conditions
Abstract
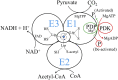
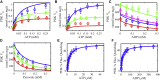
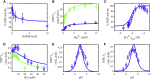
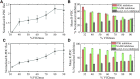
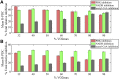
The mammalian pyruvate dehydrogenase complex (PDC) is a mitochondrial multienzyme complex that connects glycolysis to the tricarboxylic acid cycle by catalyzing pyruvate oxidation to produce acetyl-CoA, NADH, and CO2. This reaction is required to aerobically utilize glucose, a preferred metabolic fuel, and is composed of three core enzymes: pyruvate dehydrogenase (E1), dihydrolipoyl transacetylase (E2), and dihydrolipoyl dehydrogenase (E3). The pyruvate-dehydrogenase-specific kinase (PDK) and pyruvate-dehydrogenase-specific phosphatase (PDP) are considered the main control mechanism of mammalian PDC activity. However, PDK and PDP activity are allosterically regulated by several effectors fully overlapping PDC substrates and products. This collection of positive and negative feedback mechanisms confounds simple predictions of relative PDC flux, especially when all effectors are dynamically modulated during metabolic states that exist in physiologically realistic conditions, such as exercise. Here, we provide, to our knowledge, the first globally fitted, pH-dependent kinetic model of the PDC accounting for the PDC core reaction because it is regulated by PDK, PDP, metal binding equilibria, and numerous allosteric effectors. The model was used to compute PDH regulatory complex flux as a function of previously determined metabolic conditions used to simulate exercise and demonstrates increased flux with exercise. Our model reveals that PDC flux in physiological conditions is primarily inhibited by product inhibition (∼60%), mostly NADH inhibition (∼30-50%), rather than phosphorylation cycle inhibition (∼40%), but the degree to which depends on the metabolic state and PDC tissue source.
Copyright © 2021. Published by Elsevier Inc.
Figures




References
-
- Modak J., Deckwer W.D., Zeng A.P. Metabolic control analysis of eucaryotic pyruvate dehydrogenase multienzyme complex. Biotechnol. Prog. 2002;18:1157–1169. - PubMed
-
- Randle P.J., Garland P.B., Newsholme E.A. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

