Microdrop Human Immunodeficiency Virus Sequencing for Incidence and Drug Resistance Surveillance
- PMID: 33517458
- PMCID: PMC8448441
- DOI: 10.1093/infdis/jiab060
Microdrop Human Immunodeficiency Virus Sequencing for Incidence and Drug Resistance Surveillance
Abstract
Background: Precise and cost-efficient human immunodeficiency virus (HIV) incidence and drug resistance surveillances are in high demand for the advancement of the 90-90-90 "treatment for all" target.
Methods: We developed microdrop HIV sequencing for the HIV incidence and drug resistance assay (HIDA), a single-blood-draw surveillance tool for incidence and drug resistance mutation (DRM) detection. We amplified full-length HIV envelope and pol gene sequences within microdroplets, and this compartmental amplification with long-read high-throughput sequencing enabled us to recover multiple unique sequences.
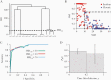
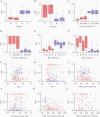
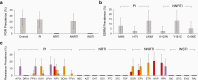
Results: We achieved greater precision in determining the stage of infection than current incidence assays, with a 1.2% false recency rate (proportion of misclassified chronic infections) and a 262-day mean duration of recent infection (average time span of recent infection classification) from 83 recently infected and 81 chronically infected individuals. Microdrop HIV sequencing demonstrated an increased capacity to detect minority variants and linked DRMs. By screening all 93 World Health Organization surveillance DRMs, we detected 6 pretreatment drug resistance mutations with 2.6%-13.2% prevalence and cross-linked mutations.
Conclusions: HIDA with microdrop HIV sequencing may promote global HIV real-time surveillance by serving as a precise and high-throughput cross-sectional survey tool that can be generalized for surveillance of other pathogens.
Keywords: Drug resistance mutations; Genomic surveillance; HIV; Incidence.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures


References
-
- UNAIDS; Global HIV & AIDS statistics — 2020 fact sheet. https://www.unaids.org/en/resources/fact-sheet Accessed 22 March 2021.
-
- World Health Organization working group on HIV incidence measurement and data use. Meeting Report. March 3-4,2018, Boston, MA, USA.:
-
- Kim AA, Rehle T. Assessing estimates of HIV incidence with a recent infection testing algorithm that includes viral load testing and exposure to antiretroviral therapy. Aids Res Hum Retrov 2018; 34:863–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

