Cell Adhesion Molecules Involved in Neurodevelopmental Pathways Implicated in 3p-Deletion Syndrome and Autism Spectrum Disorder
- PMID: 33519384
- PMCID: PMC7838543
- DOI: 10.3389/fncel.2020.611379
Cell Adhesion Molecules Involved in Neurodevelopmental Pathways Implicated in 3p-Deletion Syndrome and Autism Spectrum Disorder
Abstract
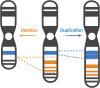
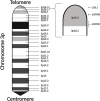
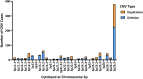
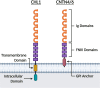
Autism spectrum disorder (ASD) is characterized by impaired social interaction, language delay and repetitive or restrictive behaviors. With increasing prevalence, ASD is currently estimated to affect 0.5-2.0% of the global population. However, its etiology remains unclear due to high genetic and phenotypic heterogeneity. Copy number variations (CNVs) are implicated in several forms of syndromic ASD and have been demonstrated to contribute toward ASD development by altering gene dosage and expression. Increasing evidence points toward the p-arm of chromosome 3 (chromosome 3p) as an ASD risk locus. Deletions occurring at chromosome 3p result in 3p-deletion syndrome (Del3p), a rare genetic disorder characterized by developmental delay, intellectual disability, facial dysmorphisms and often, ASD or ASD-associated behaviors. Therefore, we hypothesize that overlapping molecular mechanisms underlie the pathogenesis of Del3p and ASD. To investigate which genes encoded in chromosome 3p could contribute toward Del3p and ASD, we performed a comprehensive literature review and collated reports investigating the phenotypes of individuals with chromosome 3p CNVs. We observe that high frequencies of CNVs occur in the 3p26.3 region, the terminal cytoband of chromosome 3p. This suggests that CNVs disrupting genes encoded within the 3p26.3 region are likely to contribute toward the neurodevelopmental phenotypes observed in individuals affected by Del3p. The 3p26.3 region contains three consecutive genes encoding closely related neuronal immunoglobulin cell adhesion molecules (IgCAMs): Close Homolog of L1 (CHL1), Contactin-6 (CNTN6), and Contactin-4 (CNTN4). CNVs disrupting these neuronal IgCAMs may contribute toward ASD phenotypes as they have been associated with key roles in neurodevelopment. CHL1, CNTN6, and CNTN4 have been observed to promote neurogenesis and neuronal survival, and regulate neuritogenesis and synaptic function. Furthermore, there is evidence that these neuronal IgCAMs possess overlapping interactomes and participate in common signaling pathways regulating axon guidance. Notably, mouse models deficient for these neuronal IgCAMs do not display strong deficits in axonal migration or behavioral phenotypes, which is in contrast to the pronounced defects in neuritogenesis and axon guidance observed in vitro. This suggests that when CHL1, CNTN6, or CNTN4 function is disrupted by CNVs, other neuronal IgCAMs may suppress behavioral phenotypes by compensating for the loss of function.
Keywords: 3p-deletion syndrome; IgCAM; autism spectrum disorder; axon guidance; copy number variation; neurogenesis; synaptic plasticity.
Copyright © 2021 Gandawijaya, Bamford, Burbach and Oguro-Ando.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Clinical and Molecular Characterization of Two Patients with CNTN6 Copy Number Variations.Cytogenet Genome Res. 2018;156(3):144-149. doi: 10.1159/000494152. Epub 2018 Dec 4. Cytogenet Genome Res. 2018. PMID: 30508811
-
Rare recurrent copy number variations in metabotropic glutamate receptor interacting genes in children with neurodevelopmental disorders.J Neurodev Disord. 2023 Apr 29;15(1):14. doi: 10.1186/s11689-023-09483-z. J Neurodev Disord. 2023. PMID: 37120522 Free PMC article.
-
Disruption of Contactin 4 in two subjects with autism in Chinese population.Gene. 2012 Sep 1;505(2):201-5. doi: 10.1016/j.gene.2012.06.051. Epub 2012 Jun 28. Gene. 2012. PMID: 22750301
-
A current view on contactin-4, -5, and -6: Implications in neurodevelopmental disorders.Mol Cell Neurosci. 2017 Jun;81:72-83. doi: 10.1016/j.mcn.2016.12.004. Epub 2017 Jan 5. Mol Cell Neurosci. 2017. PMID: 28064060 Review.
-
[Autism spectrum disorder and genes for synaptic proteins].Brain Nerve. 2012 Jan;64(1):65-70. Brain Nerve. 2012. PMID: 22223503 Review. Japanese.
Cited by
-
Interactions of Polychlorinated Biphenyls and Their Metabolites with the Brain and Liver Transcriptome of Female Mice.ACS Chem Neurosci. 2024 Nov 6;15(21):3991-4009. doi: 10.1021/acschemneuro.4c00367. Epub 2024 Oct 11. ACS Chem Neurosci. 2024. PMID: 39392776 Free PMC article.
-
Patient Brain Organoids Identify a Link between the 16p11.2 Copy Number Variant and the RBFOX1 Gene.ACS Chem Neurosci. 2023 Nov 15;14(22):3993-4012. doi: 10.1021/acschemneuro.3c00442. Epub 2023 Oct 30. ACS Chem Neurosci. 2023. PMID: 37903506 Free PMC article.
-
Neurofilament accumulations in amyotrophic lateral sclerosis patients' motor neurons impair axonal initial segment integrity.Cell Mol Life Sci. 2023 May 15;80(6):150. doi: 10.1007/s00018-023-04797-6. Cell Mol Life Sci. 2023. PMID: 37184603 Free PMC article.
-
A study of genetic heterogeneity in autism spectrum disorders based on plasma proteomic and metabolomic analysis: multiomics study of autism heterogeneity.MedComm (2020). 2023 Sep 24;4(5):e380. doi: 10.1002/mco2.380. eCollection 2023 Oct. MedComm (2020). 2023. PMID: 37752942 Free PMC article.
-
A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease.Biomolecules. 2022 Jan 6;12(1):84. doi: 10.3390/biom12010084. Biomolecules. 2022. PMID: 35053232 Free PMC article. Review.
References
-
- American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders, 5th Edn Washington, DC: American Psychiatric Association, 10.1176/appi.books.9780890425596 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

