A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2
- PMID: 33520332
- PMCID: PMC7833526
- DOI: 10.1016/j.eng.2020.07.015
A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2
Abstract
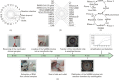
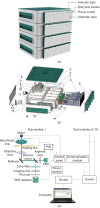
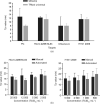
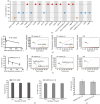
Fast and accurate diagnosis and the immediate isolation of patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are regarded as the most effective measures to restrain the coronavirus disease 2019 (COVID-19) pandemic. Here, we present a high-throughput, multi-index nucleic acid isothermal amplification analyzer (RTisochip™-W) employing a centrifugal microfluidic chip to detect 19 common respiratory viruses, including SARS-CoV-2, from 16 samples in a single run within 90 min. The limits of detection of all the viruses analyzed by the RTisochip™-W system were equal to or less than 50 copies·μL-1, which is comparable to those of conventional reverse transcription polymerase chain reaction. We also demonstrate that the RTisochip™-W system possesses the advantages of good repeatability, strong robustness, and high specificity. Finally, we analyzed 201 cases of preclinical samples, 14 cases of COVID-19-positive samples, 25 cases of clinically diagnosed samples, and 614 cases of clinical samples from patients or suspected patients with respiratory tract infections using the RTisochip™-W system. The test results matched the referenced results well and reflected the epidemic characteristics of the respiratory infectious diseases. The coincidence rate of the RTisochip™-W with the referenced kits was 98.15% for the detection of SARS-CoV-2. Based on these extensive trials, we believe that the RTisochip™-W system provides a powerful platform for fighting the COVID-19 pandemic.
Keywords: Coronavirus disease 2019; Isothermal amplification; Microfluidics; Nucleic acid testing; Severe acute respiratory syndrome coronavirus 2.
© 2020 THE AUTHORS.
Figures


Similar articles
-
Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test.PLoS One. 2021 Feb 23;16(2):e0243183. doi: 10.1371/journal.pone.0243183. eCollection 2021. PLoS One. 2021. PMID: 33621263 Free PMC article.
-
Development and Clinical Application of a Rapid and Sensitive Loop-Mediated Isothermal Amplification Test for SARS-CoV-2 Infection.mSphere. 2020 Aug 26;5(4):e00808-20. doi: 10.1128/mSphere.00808-20. mSphere. 2020. PMID: 32848011 Free PMC article.
-
Developing a Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Seven Respiratory Viruses including SARS-CoV-2.Medicina (Kaunas). 2022 Sep 5;58(9):1224. doi: 10.3390/medicina58091224. Medicina (Kaunas). 2022. PMID: 36143901 Free PMC article.
-
Toward a next-generation diagnostic tool: A review on emerging isothermal nucleic acid amplification techniques for the detection of SARS-CoV-2 and other infectious viruses.Anal Chim Acta. 2022 May 29;1209:339338. doi: 10.1016/j.aca.2021.339338. Epub 2021 Dec 1. Anal Chim Acta. 2022. PMID: 35569864 Free PMC article. Review.
-
[Isothermal amplification technology based on microfluidic chip].Sheng Wu Gong Cheng Xue Bao. 2022 Mar 25;38(3):943-960. doi: 10.13345/j.cjb.210533. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 35355466 Review. Chinese.
Cited by
-
Microfluidic-based virus detection methods for respiratory diseases.Emergent Mater. 2021;4(1):143-168. doi: 10.1007/s42247-021-00169-7. Epub 2021 Mar 25. Emergent Mater. 2021. PMID: 33786415 Free PMC article. Review.
-
A precise review on NAATs-based diagnostic assays for COVID-19: A motion in fast POC molecular tests.Eur J Clin Invest. 2022 Nov;52(11):e13853. doi: 10.1111/eci.13853. Epub 2022 Aug 30. Eur J Clin Invest. 2022. PMID: 35989561 Free PMC article. Review.
-
Detection of Multiplex NASBA RNA Products Using Colorimetric Split G Quadruplex Probes.Methods Mol Biol. 2023;2709:287-298. doi: 10.1007/978-1-0716-3417-2_20. Methods Mol Biol. 2023. PMID: 37572289
-
Multiplexed electrochemical liposomes applied to the detection of nucleic acids for Influenza A, Influenza B and SARS-CoV-2.Anal Bioanal Chem. 2024 Jun;416(15):3487-3500. doi: 10.1007/s00216-024-05145-8. Epub 2024 Jan 19. Anal Bioanal Chem. 2024. PMID: 38240795 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
