Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer
- PMID: 33521652
- PMCID: PMC7842382
- DOI: 10.1038/s43018-019-0020-z
Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer
Abstract
The natural history of small cell lung cancer (SCLC) includes rapid evolution from chemosensitivity to chemoresistance, although mechanisms underlying this evolution remain obscure due to scarcity of post-relapse tissue samples. We generated circulating tumor cell (CTC)-derived xenografts (CDXs) from SCLC patients to study intratumoral heterogeneity (ITH) via single-cell RNAseq of chemo-sensitive and -resistant CDXs and patient CTCs. We found globally increased ITH including heterogeneous expression of therapeutic targets and potential resistance pathways, such as EMT, between cellular subpopulations following treatment-resistance. Similarly, serial profiling of patient CTCs directly from blood confirmed increased ITH post-relapse. These data suggest that treatment-resistance in SCLC is characterized by coexisting subpopulations of cells with heterogeneous gene expression leading to multiple, concurrent resistance mechanisms. These findings emphasize the need for clinical efforts to focus on rational combination therapies for treatment-naïve SCLC tumors to maximize initial responses and counteract the emergence of ITH and diverse resistance mechanisms.
Keywords: CDX; CTC; SCLC; intratumoral heterogeneity; single-cell RNAseq.
Conflict of interest statement
COMPETING INTERESTS STATEMENT L.A.B. serves on advisory committees for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar SA, Sierra Oncology, Merck, Bristol Myers Squibb, Genentech, and Pfizer and has research support from AbbVie, AstraZeneca, GenMab, Sierra Oncology, Tolero Pharmaceuticals. J.V.H. serves on advisory committees for AstraZeneca, Boehringer Ingelheim, Exelixis, Genentech, GSK, Guardant Health, Hengrui, Lilly, Novartis, Spectrum, EMD Serono, and Synta, has research support from AstraZeneca, Bayer, GlaxoSmithKline, and Spectrum and royalties and licensing fees from Spectrum. Otherwise, there are no pertinent financial or non-financial conflicts of interest to report.
Figures





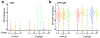




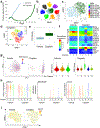

Comment in
-
The therapy escapes of small-cell lung cancer.Nat Cancer. 2020 Apr;1(4):374-375. doi: 10.1038/s43018-020-0058-y. Nat Cancer. 2020. PMID: 35121967 No abstract available.
References
-
- in H. R. 733 (ed Senate and House of Representatives of the United States of America in Congress assembled) (2012).
-
- Drapkin BJ et al. Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer discovery, doi: 10.1158/2159-8290.CD-17-0935 (2018). - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

