A distinct innate immune signature marks progression from mild to severe COVID-19
- PMID: 33521697
- PMCID: PMC7817872
- DOI: 10.1016/j.xcrm.2020.100166
A distinct innate immune signature marks progression from mild to severe COVID-19
Abstract
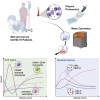
Coronavirus disease 2019 (COVID-19) manifests with a range of severities, but immune signatures of mild and severe disease are still not fully understood. Here, we use mass cytometry and targeted proteomics to profile the innate immune response of patients with mild or severe COVID-19 and of healthy individuals. Sampling at different stages allows us to reconstruct a pseudo-temporal trajectory of the innate response. A surge of CD169+ monocytes associated with an IFN-γ+MCP-2+ signature rapidly follows symptom onset. At later stages, we observe a persistent inflammatory phenotype in patients with severe disease, dominated by high CCL3 and CCL4 abundance correlating with the re-appearance of CD16+ monocytes, whereas the response of mild COVID-19 patients normalizes. Our data provide insights into the dynamic nature of inflammatory responses in COVID-19 patients and identify sustained innate immune responses as a likely mechanism in severe patients, thus supporting the investigation of targeted interventions in severe COVID-19.
Keywords: CCL3; CD169; COVID-19; MIP1α; SARS-CoV-2; emergency granulopoiesis; inflammation; innate immune response; mass cytometry; monocytes; proteomics; sialoadhesin.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. - PubMed
-
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

