Targeted brachyury degradation disrupts a highly specific autoregulatory program controlling chordoma cell identity
- PMID: 33521702
- PMCID: PMC7817874
- DOI: 10.1016/j.xcrm.2020.100188
Targeted brachyury degradation disrupts a highly specific autoregulatory program controlling chordoma cell identity
Abstract
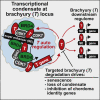
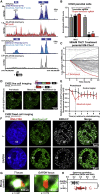
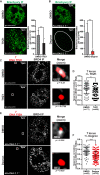
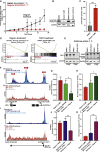
Chordomas are rare spinal tumors addicted to expression of the developmental transcription factor brachyury. In chordomas, brachyury is super-enhancer associated and preferentially downregulated by pharmacologic transcriptional CDK inhibition, leading to cell death. To understand the underlying basis of this sensitivity, we dissect the brachyury transcription regulatory network and compare the consequences of brachyury degradation with transcriptional CDK inhibition. Brachyury defines the chordoma super-enhancer landscape and autoregulates through binding its super-enhancer, and its locus forms a transcriptional condensate. Transcriptional CDK inhibition and brachyury degradation disrupt brachyury autoregulation, leading to loss of its transcriptional condensate and transcriptional program. Compared with transcriptional CDK inhibition, which globally downregulates transcription, leading to cell death, brachyury degradation is much more selective, inducing senescence and sensitizing cells to anti-apoptotic inhibition. These data suggest that brachyury downregulation is a core tenet of transcriptional CDK inhibition and motivates developing strategies to target brachyury and its autoregulatory feedback loop.
Keywords: brachyury; chordoma; cyclin-dependent kinase; phase separation; super-enhancer; targeted degradation; transcription; transcription factor; transcriptional condensate; transcriptional inhibition.
© 2020 The Authors.
Conflict of interest statement
C.Y.L. is an inventor of intellectual property licensed to Syros Pharmaceuticals, and is the Vice President of Biology and a shareholder of Kronos Bio, Inc. S.L.S. is a shareholder and serves on the Board of Directors of Jnana Therapeutics; is a shareholder of Forma Therapeutics and Decibel Therapeutics; is a shareholder and advises Kojin Therapeutics, Kisbee Therapeutics, Exo Therapeutics, and Eikonizo Therapeutics; serves on the Scientific Advisory Boards of Eisai Co., Ltd., Ono Pharma Foundation, Biogen, Inc., and F-Prime Capital Partners and the Board of Advisers of the Genomics Institute of the Novartis Research Foundation; and is a Novartis Faculty Scholar. M.J.W. is an employee and equity holder of Kojin Therapeutics. P.A.C. is an advisor to Pfizer, Inc. and nference, Inc. R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. P.W. is a current employee of The Institute of Cancer Research, which has a Rewards to Inventors scheme and has a commercial interest in development of inhibitors of the WNT pathway, CDK8/19, and other CDKs, with intellectual property licensed to Merck and Cyclacel Pharmaceuticals. P.W. is also a consultant for Astex Pharmaceuticals, CV6 Therapeutics, Nextechinvest, Nuevolution, Black Diamond, and STORM Therapeutics and holds equity in Chroma Therapeutics, Nextech, Black Diamond, and STORM. B.N. is an inventor on patent applications related to the dTAG system (WO/2017/024318, WO/2017/024319, WO/2018/148443, and WO/2018/148440). N.S.G. is a Scientific Founder, member of the Scientific Advisory Board (SAB), and equity holder in C4 Therapeutics, Syros, Soltego (board member), B2S, Allorion, and Inception. The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voroni, Arbella, Deerfield, and Sanofi.
Figures



References
-
- Gonda T.J., Ramsay R.G. Directly targeting transcriptional dysregulation in cancer. Nat. Rev. Cancer. 2015;15:686–694. - PubMed
-
- Stewart E., McEvoy J., Wang H., Chen X., Honnell V., Ocarz M., Gordon B., Dapper J., Blankenship K., Yang Y., St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell. 2018;34:411–426.e19. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

