Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study
- PMID: 33521709
- PMCID: PMC7837364
- DOI: 10.1016/S2666-5247(20)30197-X
Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study
Abstract
Background: Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces an antibody response targeting multiple antigens that changes over time. This study aims to take advantage of this complexity to develop more accurate serological diagnostics.
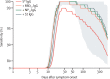
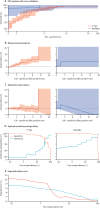
Methods: A multiplex serological assay was developed to measure IgG and IgM antibody responses to seven SARS-CoV-2 spike or nucleoprotein antigens, two antigens for the nucleoproteins of the 229E and NL63 seasonal coronaviruses, and three non-coronavirus antigens. Antibodies were measured in serum samples collected up to 39 days after symptom onset from 215 adults in four French hospitals (53 patients and 162 health-care workers) with quantitative RT-PCR-confirmed SARS-CoV-2 infection, and negative control serum samples collected from healthy adult blood donors before the start of the SARS-CoV-2 epidemic (335 samples from France, Thailand, and Peru). Machine learning classifiers were trained with the multiplex data to classify individuals with previous SARS-CoV-2 infection, with the best classification performance displayed by a random forests algorithm. A Bayesian mathematical model of antibody kinetics informed by prior information from other coronaviruses was used to estimate time-varying antibody responses and assess the sensitivity and classification performance of serological diagnostics during the first year following symptom onset. A statistical estimator is presented that can provide estimates of seroprevalence in very low-transmission settings.
Findings: IgG antibody responses to trimeric spike protein (Stri) identified individuals with previous SARS-CoV-2 infection with 91·6% (95% CI 87·5-94·5) sensitivity and 99·1% (97·4-99·7) specificity. Using a serological signature of IgG and IgM to multiple antigens, it was possible to identify infected individuals with 98·8% (96·5-99·6) sensitivity and 99·3% (97·6-99·8) specificity. Informed by existing data from other coronaviruses, we estimate that 1 year after infection, a monoplex assay with optimal anti-Stri IgG cutoff has 88·7% (95% credible interval 63·4-97·4) sensitivity and that a four-antigen multiplex assay can increase sensitivity to 96·4% (80·9-100·0). When applied to population-level serological surveys, statistical analysis of multiplex data allows estimation of seroprevalence levels less than 2%, below the false-positivity rate of many other assays.
Interpretation: Serological signatures based on antibody responses to multiple antigens can provide accurate and robust serological classification of individuals with previous SARS-CoV-2 infection. This provides potential solutions to two pressing challenges for SARS-CoV-2 serological surveillance: classifying individuals who were infected more than 6 months ago and measuring seroprevalence in serological surveys in very low-transmission settings.
Funding: European Research Council. Fondation pour la Recherche Médicale. Institut Pasteur Task Force COVID-19.
© 2021 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license.
Figures


References
-
- The Foundation for Innovative New Diagnostics SARS-CoV-2 diagnostics: performance data. https://www.finddx.org/covid-19/dx-data
-
- White MT, Griffin JT, Akpogheneta O. Dynamics of the antibody response to Plasmodium falciparum infection in African children. J Infect Dis. 2014;210:1115–1122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

