Harnessing machine learning for development of microbiome therapeutics
- PMID: 33522391
- PMCID: PMC7872042
- DOI: 10.1080/19490976.2021.1872323
Harnessing machine learning for development of microbiome therapeutics
Abstract
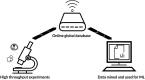
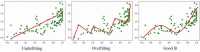
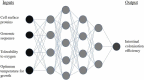
The last twenty years of seminal microbiome research has uncovered microbiota's intrinsic relationship with human health. Studies elucidating the relationship between an unbalanced microbiome and disease are currently published daily. As such, microbiome big data have become a reality that provide a mine of information for the development of new therapeutics. Machine learning (ML), a branch of artificial intelligence, offers powerful techniques for big data analysis and prediction-making, that are out of reach of human intellect alone. This review will explore how ML can be applied for the development of microbiome-targeted therapeutics. A background on ML will be given, followed by a guide on where to find reliable microbiome big data. Existing applications and opportunities will be discussed, including the use of ML to discover, design, and characterize microbiome therapeutics. The use of ML to optimize advanced processes, such as 3D printing and in silico prediction of drug-microbiome interactions, will also be highlighted. Finally, barriers to adoption of ML in academic and industrial settings will be examined, concluded by a future outlook for the field.
Keywords: COVID-19; artificial intelligence; clinical translation; colonic drug delivery; drug product development; machine learning; microbial therapeutics; microbiome; personalized medicines; pharmaceutical sciences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
