Post-transcriptional regulation of redox homeostasis by the small RNA SHOxi in haloarchaea
- PMID: 33522404
- PMCID: PMC8583180
- DOI: 10.1080/15476286.2021.1874717
Post-transcriptional regulation of redox homeostasis by the small RNA SHOxi in haloarchaea
Abstract
While haloarchaea are highly resistant to oxidative stress, a comprehensive understanding of the processes regulating this remarkable response is lacking. Oxidative stress-responsive small non-coding RNAs (sRNAs) have been reported in the model archaeon, Haloferax volc anii, but targets and mechanisms have not been elucidated. Using a combination of high throughput and reverse molecular genetic approaches, we elucidated the functional role of the most up-regulated intergenic sRNA during oxidative stress in H. volcanii, named
Keywords: Non-coding RNA; archaea; gene regulation; oxidative stress; redox homoeostasis; small RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






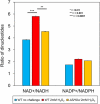

References
-
- Cech TR, Steitz JA.. The noncoding RNA revolution: trashing old rules to forge new ones. Cell. 2014;157(1):77–94. - PubMed
-
- Wagner, E.G.H.; Romby, P. Chapter Three—Small RNAs in Bacteria and Archaea: Who They Are, What They Do and How They Do It. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Academic Press: Cambridge, MA, USA, 2015. pp. 133–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
