Identification of genes associated with the biosynthesis of unsaturated fatty acid and oil accumulation in herbaceous peony 'Hangshao' (Paeonia lactiflora 'Hangshao') seeds based on transcriptome analysis
- PMID: 33522906
- PMCID: PMC7849092
- DOI: 10.1186/s12864-020-07339-7
Identification of genes associated with the biosynthesis of unsaturated fatty acid and oil accumulation in herbaceous peony 'Hangshao' (Paeonia lactiflora 'Hangshao') seeds based on transcriptome analysis
Abstract
Background: Paeonia lactiflora 'Hangshao' is widely cultivated in China as a traditional Chinese medicine 'Radix Paeoniae Alba'. Due to the abundant unsaturated fatty acids in its seed, it can also be regarded as a new oilseed plant. However, the process of the biosynthesis of unsaturated fatty acids in it has remained unknown. Therefore, transcriptome analysis is helpful to better understand the underlying molecular mechanisms.
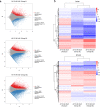
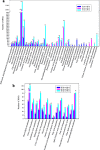
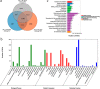
Results: Five main fatty acids were detected, including stearic acid, palmitic acid, oleic acid, linoleic acid and α-linolenic acid, and their absolute contents first increased and then decreased during seed development. A total of 150,156 unigenes were obtained by transcriptome sequencing. There were 15,005 unigenes annotated in the seven functional databases, including NR, NT, GO, KOG, KEGG, Swiss-Prot and InterPro. Based on the KEGG database, 1766 unigenes were annotated in the lipid metabolism. There were 4635, 12,304, and 18,291 DEGs in Group I (60 vs 30 DAF), Group II (90 vs 60 DAF) and Group III (90 vs 30 DAF), respectively. A total of 1480 DEGs were detected in the intersection of the three groups. In 14 KEGG pathways of lipid metabolism, 503 DEGs were found, belonging to 111 enzymes. We screened out 123 DEGs involved in fatty acid biosynthesis (39 DEGs), fatty acid elongation (33 DEGs), biosynthesis of unsaturated fatty acid (24 DEGs), TAG assembly (17 DEGs) and lipid storage (10 DEGs). Furthermore, qRT-PCR was used to analyze the expression patterns of 16 genes, including BBCP, BC, MCAT, KASIII, KASII, FATA, FATB, KCR, SAD, FAD2, FAD3, FAD7, GPAT, DGAT, OLE and CLO, most of which showed the highest expression at 45 DAF, except for DGAT, OLE and CLO, which showed the highest expression at 75 DAF.
Conclusions: We predicted that MCAT, KASIII, FATA, SAD, FAD2, FAD3, DGAT and OLE were the key genes in the unsaturated fatty acid biosynthesis and oil accumulation in herbaceous peony seed. This study provides the first comprehensive genomic resources characterizing herbaceous peony seed gene expression at the transcriptional level. These data lay the foundation for elucidating the molecular mechanisms of fatty acid biosynthesis and oil accumulation for herbaceous peony.
Keywords: Differentially expressed gene; Fatty acid biosynthesis; Paeonia lactiflora ‘Hangshao’; Transcriptome; qRT-PCR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Qing KJ. Illustrtion of one hundred ornamental flowers bonsai-the herbaceous peony. Beijing: China Forestry Press; 2004.
-
- Zhu MX, Li SN, You HD, Han B, Wang ZP, Hu YX, et al. Simultaneous determination of paeoniflorin and albiflorin in radix paeoniae Rubra by HPLC–DAD–ELSD. Acta Chromatogr. 2017;29(2):279–289. doi: 10.1556/1326.2017.29.2.12. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

