Factors influencing estimates of HIV-1 infection timing using BEAST
- PMID: 33524022
- PMCID: PMC7877758
- DOI: 10.1371/journal.pcbi.1008537
Factors influencing estimates of HIV-1 infection timing using BEAST
Abstract
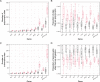
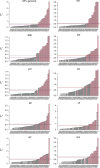
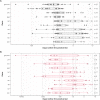
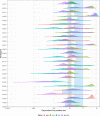
While large datasets of HIV-1 sequences are increasingly being generated, many studies rely on a single gene or fragment of the genome and few comparative studies across genes have been done. We performed genome-based and gene-specific Bayesian phylogenetic analyses to investigate how certain factors impact estimates of the infection dates in an acute HIV-1 infection cohort, RV217. In this cohort, HIV-1 diagnosis corresponded to the first RNA positive test and occurred a median of four days after the last negative test, allowing us to compare timing estimates using BEAST to a narrow window of infection. We analyzed HIV-1 sequences sampled one week, one month and six months after HIV-1 diagnosis in 39 individuals. We found that shared diversity and temporal signal was limited in acute infection, and insufficient to allow timing inferences in the shortest HIV-1 genes, thus dated phylogenies were primarily analyzed for env, gag, pol and near full-length genomes. There was no one best-fitting model across participants and genes, though relaxed molecular clocks (73% of best-fitting models) and the Bayesian skyline (49%) tended to be favored. For infections with single founders, the infection date was estimated to be around one week pre-diagnosis for env (IQR: 3-9 days) and gag (IQR: 5-9 days), whilst the genome placed it at a median of 10 days (IQR: 4-19). Multiply-founded infections proved problematic to date. Our ability to compare timing inferences to precise estimates of HIV-1 infection (within a week) highlights that molecular dating methods can be applied to within-host datasets from early infection. Nonetheless, our results also suggest caution when using uniform clock and population models or short genes with limited information content.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Delaney KP, Hanson DL, Masciotra S, Ethridge SF, Wesolowski L, Owen SM. Time Until Emergence of HIV Test Reactivity Following Infection With HIV-1: Implications for Interpreting Test Results and Retesting After Exposure. Clinical infectious diseases. 2017;64: 53–59. 10.1093/cid/ciw666 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

