Archaeal tyrosine recombinases
- PMID: 33524101
- PMCID: PMC8371274
- DOI: 10.1093/femsre/fuab004
Archaeal tyrosine recombinases
Abstract
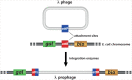
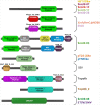
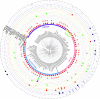
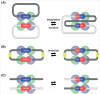
The integration of mobile genetic elements into their host chromosome influences the immediate fate of cellular organisms and gradually shapes their evolution. Site-specific recombinases catalyzing this integration have been extensively characterized both in bacteria and eukarya. More recently, a number of reports provided the in-depth characterization of archaeal tyrosine recombinases and highlighted new particular features not observed in the other two domains. In addition to being active in extreme environments, archaeal integrases catalyze reactions beyond site-specific recombination. Some of these integrases can catalyze low-sequence specificity recombination reactions with the same outcome as homologous recombination events generating deep rearrangements of their host genome. A large proportion of archaeal integrases are termed suicidal due to the presence of a specific recombination target within their own gene. The paradoxical maintenance of integrases that disrupt their gene upon integration implies novel mechanisms for their evolution. In this review, we assess the diversity of the archaeal tyrosine recombinases using a phylogenomic analysis based on an exhaustive similarity network. We outline the biochemical, ecological and evolutionary properties of these enzymes in the context of the families we identified and emphasize similarities and differences between archaeal recombinases and their bacterial and eukaryal counterparts.
Keywords: Archaea; genome evolution; horizontal transfer; mobile genetic element; site-specific recombination; tyrosine recombinase.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Figures








Similar articles
-
Pervasive Suicidal Integrases in Deep-Sea Archaea.Mol Biol Evol. 2020 Jun 1;37(6):1727-1743. doi: 10.1093/molbev/msaa041. Mol Biol Evol. 2020. PMID: 32068866 Free PMC article.
-
Site-Specific Recombination by SSV2 Integrase: Substrate Requirement and Domain Functions.J Virol. 2015 Nov;89(21):10934-44. doi: 10.1128/JVI.01637-15. Epub 2015 Aug 19. J Virol. 2015. PMID: 26292330 Free PMC article.
-
A novel family of tyrosine integrases encoded by the temperate pleolipovirus SNJ2.Nucleic Acids Res. 2018 Mar 16;46(5):2521-2536. doi: 10.1093/nar/gky005. Nucleic Acids Res. 2018. PMID: 29361162 Free PMC article.
-
Systematic Discovery of a New Catalogue of Tyrosine-Type Integrases in Bacterial Genomic Islands.Appl Environ Microbiol. 2023 Feb 28;89(2):e0173822. doi: 10.1128/aem.01738-22. Epub 2023 Jan 31. Appl Environ Microbiol. 2023. PMID: 36719242 Free PMC article.
-
Archaeal integrative genetic elements and their impact on genome evolution.Res Microbiol. 2002 Jul-Aug;153(6):325-32. doi: 10.1016/s0923-2508(02)01331-1. Res Microbiol. 2002. PMID: 12234006 Review.
Cited by
-
How and when organisms edit their own genomes.Nat Genet. 2025 Aug;57(8):1823-1834. doi: 10.1038/s41588-025-02230-1. Epub 2025 Jun 27. Nat Genet. 2025. PMID: 40579538 Review.
-
Systematic identification of cargo-mobilizing genetic elements reveals new dimensions of eukaryotic diversity.Nucleic Acids Res. 2024 Jun 10;52(10):5496-5513. doi: 10.1093/nar/gkae327. Nucleic Acids Res. 2024. PMID: 38686785 Free PMC article.
-
Discovery and characterization of complete genomes of 38 head-tailed proviruses in four predominant phyla of archaea.Microbiol Spectr. 2025 Jan 7;13(1):e0049224. doi: 10.1128/spectrum.00492-24. Epub 2024 Nov 15. Microbiol Spectr. 2025. PMID: 39545734 Free PMC article.
-
Provirus deletion from Haloferax volcanii affects motility, stress resistance, and CRISPR RNA expression.Microlife. 2025 May 19;6:uqaf008. doi: 10.1093/femsml/uqaf008. eCollection 2025. Microlife. 2025. PMID: 40395997 Free PMC article.
-
Viruses of the Turriviridae: an emerging model system for studying archaeal virus-host interactions.Front Microbiol. 2023 Sep 21;14:1258997. doi: 10.3389/fmicb.2023.1258997. eCollection 2023. Front Microbiol. 2023. PMID: 37808280 Free PMC article. Review.
References
-
- Abremski K, Gottesman S. Purification of the bacteriophage lambda Xis gene product required for lambda excisive recombination. J Biol Chem. 1982;257:9658–62. - PubMed
-
- Abremski K, Wierzbicki A, Frommer Bet al. . Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J Biol Chem. 1986;261:391–6. - PubMed
-
- Abremski KE, Hoess RH. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992;5:87–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

