Doxorubicin-induced senescence promotes stemness and tumorigenicity in EpCAM-/CD133- nonstem cell population in hepatocellular carcinoma cell line, HuH-7
- PMID: 33524223
- PMCID: PMC8334288
- DOI: 10.1002/1878-0261.12916
Doxorubicin-induced senescence promotes stemness and tumorigenicity in EpCAM-/CD133- nonstem cell population in hepatocellular carcinoma cell line, HuH-7
Abstract
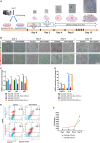
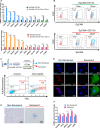
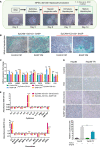
The therapeutic induction of senescence is a potential means to treat cancer, primarily acting through the induction of a persistent growth-arrested state in tumors. However, recent studies have indicated that therapy-induced senescence (TIS) in tumor cells allows for the prolonged survival of a subgroup of cells in a dormant state, with the potential to re-enter the cell cycle along with an increased stemness gene expression. Residual cells after TIS with increased cancer stem cell phenotype may have profound implications for tumor aggressiveness and disease recurrence. Herein, we investigated senescence-associated stemness in EpCAM+/CD133+ liver cancer stem cell and EpCAM-/CD133- nonstem cell populations in HuH7 cell line. We demonstrated that treatment with doxorubicin induces senescence in both cell populations, accompanied by a significant increase in the expression of reprogramming genes SOX2, KLF4, and c-MYC as well as liver stemness-related genes EpCAM, CK19, and ANXA3 and the multidrug resistance-related gene ABCG2. Moreover, doxorubicin treatment significantly increased EpCAM + population in nonstem cells indicating senescence-associated reprogramming of nonstem cell population. Also, Wnt/β-catenin target genes were increased in these cells, while inhibition of this signaling pathway decreased stem cell gene expression. Importantly, Dox-treated EpCAM-/CD133- nonstem cells had increased in vivo tumor-forming ability. In addition, when SASP-CM from Dox-treated cells were applied onto hİPSC-derived hepatocytes, senescence was induced in hepatocytes along with an increased expression of TGF-β, KLF4, and AXIN2. Importantly, SASP-CM was not able to induce senescence in Hep3B-TR cells, a derivative line rendered resistant to TGF-β signaling. Furthermore, ELISA experiments revealed that the SASP-CM of Dox-treated cells contain inflammatory cytokines IL8 and IP10. In summary, our findings further emphasize the importance of carefully dissecting the beneficial and detrimental aspects of prosenescence therapy in HCC and support the potential use of senolytic drugs in HCC treatment in order to eliminate adverse effects of TIS.
Keywords: EpCAM; WNT; cancer stem cell; hepatocellular carcinoma; therapy-induced senescence.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Friemel J, Rechsteiner M, Frick L, Böhm F, Struckmann K, Egger M, Moch H, Heikenwalder M & Weber A (2015) Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 21, 1951–1961. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

