Mucosal Genomics Implicate Lymphocyte Activation and Lipid Metabolism in Refractory Environmental Enteric Dysfunction
- PMID: 33524399
- PMCID: PMC8113748
- DOI: 10.1053/j.gastro.2021.01.221
Mucosal Genomics Implicate Lymphocyte Activation and Lipid Metabolism in Refractory Environmental Enteric Dysfunction
Abstract
Background & aims: Environmental enteric dysfunction (EED) limits the Sustainable Development Goals of improved childhood growth and survival. We applied mucosal genomics to advance our understanding of EED.
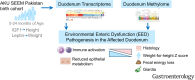
Methods: The Study of Environmental Enteropathy and Malnutrition (SEEM) followed 416 children from birth to 24 months in a rural district in Pakistan. Biomarkers were measured at 9 months and tested for association with growth at 24 months. The duodenal methylome and transcriptome were determined in 52 undernourished SEEM participants and 42 North American controls and patients with celiac disease.
Results: After accounting for growth at study entry, circulating insulin-like growth factor-1 (IGF-1) and ferritin predicted linear growth, whereas leptin correlated with future weight gain. The EED transcriptome exhibited suppression of antioxidant, detoxification, and lipid metabolism genes, and induction of anti-microbial response, interferon, and lymphocyte activation genes. Relative to celiac disease, suppression of antioxidant and detoxification genes and induction of antimicrobial response genes were EED-specific. At the epigenetic level, EED showed hyper-methylation of epithelial metabolism and barrier function genes, and hypo-methylation of immune response and cell proliferation genes. Duodenal coexpression modules showed association between lymphocyte proliferation and epithelial metabolic genes and histologic severity, fecal energy loss, and wasting (weight-for-length/height Z < -2.0). Leptin was associated with expression of epithelial carbohydrate metabolism and stem cell renewal genes. Immune response genes were attenuated by giardia colonization.
Conclusions: Children with reduced circulating IGF-1 are more likely to experience stunting. Leptin and a gene signature for lymphocyte activation and dysregulated lipid metabolism are implicated in wasting, suggesting new approaches for EED refractory to nutritional intervention. ClinicalTrials.gov, Number: NCT03588013. (https://clinicaltrials.gov/ct2/show/NCT03588013).
Keywords: Anthropometrics; DNA Methylation; Intestine; RNA Sequencing.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Lunn P.G., Northrop-Clewes C.A., Downes R.M. Intestinal permeability, mucosal injury, and growth faltering in Gambian infants. Lancet. 1991;338:907–910. - PubMed
-
- Fasano A., Catassi C. Clinical practice. Celiac disease. N Engl J Med. 2012;367:2419–2426. - PubMed
-
- Campbell D.I., Murch S.H., Elia M. Chronic T cell-mediated enteropathy in rural west African children: relationship with nutritional status and small bowel function. Pediatr Res. 2003;54:306–311. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

