PROTACs and Building Blocks: The 2D Chemical Space in Very Early Drug Discovery
- PMID: 33525371
- PMCID: PMC7865272
- DOI: 10.3390/molecules26030672
PROTACs and Building Blocks: The 2D Chemical Space in Very Early Drug Discovery
Abstract
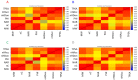
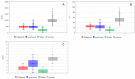
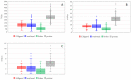
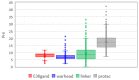
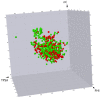
Targeted protein degradation by PROTACs has emerged as a new modality for the knockdown of a range of proteins, and, more recently, it has become increasingly clear that the PROTAC chemical space requires characterization through a pool of ad hoc physicochemical descriptors. In this study, a new database named PROTAC-DB that provides extensive information about PROTACs and building blocks was used to obtain the 2D chemical structures of about 1600 PROTACs, 60 E3 ligands, 800 linkers, and 202 warheads. For every structure, we calculated a pool of seven 2D descriptors carefully identified as informative for large and flexible structures. For comparison purposes, the same procedure was applied to a dataset of about 50 bRo5 approved drugs reported in the literature. Correlation matrices, PCAs, box plots, and other graphical tools were used to define and understand the chemical space covered by PROTACs and building blocks in relation to other compounds. Results show that linkers have different properties than E3 ligands and warheads. Polar descriptors additivity is not respected when passing from building blocks to degraders. Moreover, a very preliminary analysis based on three PROTACs with high, intermediate, and low permeability showed how the most permeable compounds seem to occupy a region closer to bRo5 drugs and, thus, exhibit different properties than impermeable compounds. Finally, a second database, PROTACpedia, was used to discuss the relevance of physicochemical descriptors on degradation activity.
Keywords: 2D physicochemical descriptors; PROTAC; chemical space; degrader; permeability.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

