Investigating the Diversity and Host Range of Novel Parvoviruses from North American Ducks Using Epidemiology, Phylogenetics, Genome Structure, and Codon Usage Analysis
- PMID: 33525386
- PMCID: PMC7912424
- DOI: 10.3390/v13020193
Investigating the Diversity and Host Range of Novel Parvoviruses from North American Ducks Using Epidemiology, Phylogenetics, Genome Structure, and Codon Usage Analysis
Abstract
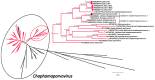
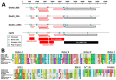
Parvoviruses are small single-stranded DNA viruses that can infect both vertebrates and invertebrates. We report here the full characterization of novel viruses we identified in ducks, including two viral species within the subfamily Hamaparvovirinae (duck-associated chapparvovirus, DAC) and a novel species within the subfamily Densovirinae (duck-associated ambidensovirus, DAAD). Overall, 5.7% and 21.1% of the 123 screened ducks (American black ducks, mallards, northern pintail) were positive for DAC and DAAD, respectively, and both viruses were more frequently detected in autumn than in winter. Genome organization and predicted transcription profiles of DAC and DAAD were similar to viruses of the genera Chaphamaparvovirus and Protoambidensovirus, respectively. Their association to these genera was also demonstrated by subfamily-wide phylogenetic and distance analyses of non-structural protein NS1 sequences. While DACs were included in a highly supported clade of avian viruses, no definitive conclusions could be drawn about the host type of DAAD because it was phylogenetically close to viruses found in vertebrates and invertebrates and analyses of codon usage bias and nucleotide frequencies of viruses within the family Parvoviridae showed no clear host-based viral segregation. This study highlights the high parvoviral diversity in the avian reservoir with many avian-associated parvoviruses likely yet to be discovered.
Keywords: avian viruses; chaphamaparvovirus; codon usage; densovirus; dinucleotide frequencies; insect viruses; parvovirus; virus discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Pénzes J.J., Söderlund-Venermo M., Canuti M., Eis-Hübinger A.M., Hughes J., Cotmore S.F., Harrach B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020;165:2133–2146. doi: 10.1007/s00705-020-04632-4. - DOI - PubMed
-
- Pénzes J.J., Canuti M., Söderlund-Venermo M., Eis-Huebinger A.M., Ogliastro M., Harrach B. ICTV Taxonomy Proposal 2020. International Committee on Taxonomy of Viruses (ICTV); 2020. Create three new genera and 19 new species (Piccovirales: Parvoviridae)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

