Angiogenin (ANG)-Ribonuclease Inhibitor (RNH1) System in Protein Synthesis and Disease
- PMID: 33525475
- PMCID: PMC7866052
- DOI: 10.3390/ijms22031287
Angiogenin (ANG)-Ribonuclease Inhibitor (RNH1) System in Protein Synthesis and Disease
Abstract
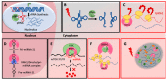
Protein synthesis is a highly complex process executed by well-organized translation machinery. Ribosomes, tRNAs and mRNAs are the principal components of this machinery whereas RNA binding proteins and ribosome interacting partners act as accessory factors. Angiogenin (ANG)-Ribonuclease inhibitor (RNH1) system is one such accessory part of the translation machinery that came into focus afresh due to its unconventional role in the translation. ANG is conventionally known for its ability to induce blood vessel formation and RNH1 as a "sentry" to protect RNAs from extracellular RNases. However, recent studies suggest them to be important in translation regulation. During cell homeostasis, ANG in the nucleus promotes rRNA transcription. While under stress, ANG translocates to the cytosol and cleaves tRNA into fragments which inhibit ribosome biogenesis and protein synthesis. RNH1, which intimately interacts with ANG to inhibit its ribonucleolytic activity, can also bind to the 40S ribosomes and control translation by yet to be known mechanisms. Here, we review recent advancement in the knowledge of translation regulation by the ANG-RNH1 system. We also gather information about this system in cell homeostasis as well as in pathological conditions such as cancer and ribosomopathies. Additionally, we discuss the future research directions and therapeutic potential of this system.
Keywords: Angiogenin (ANG); RNases; Ribonuclease inhibitor (RNH1); mRNA translation regulation; ribosomal heterogeneity; transcript-specific translation and ribosomopathies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

