Phylodynamics and Codon Usage Pattern Analysis of Broad Bean Wilt Virus 2
- PMID: 33525612
- PMCID: PMC7912035
- DOI: 10.3390/v13020198
Phylodynamics and Codon Usage Pattern Analysis of Broad Bean Wilt Virus 2
Abstract
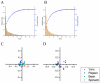
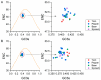
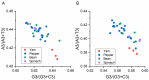
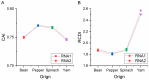
Broad bean wilt virus 2 (BBWV-2), which belongs to the genus Fabavirus of the family Secoviridae, is an important pathogen that causes damage to broad bean, pepper, yam, spinach and other economically important ornamental and horticultural crops worldwide. Previously, only limited reports have shown the genetic variation of BBWV2. Meanwhile, the detailed evolutionary changes, synonymous codon usage bias and host adaptation of this virus are largely unclear. Here, we performed comprehensive analyses of the phylodynamics, reassortment, composition bias and codon usage pattern of BBWV2 using forty-two complete genome sequences of BBWV-2 isolates together with two other full-length RNA1 sequences and six full-length RNA2 sequences. Both recombination and reassortment had a significant influence on the genomic evolution of BBWV2. Through phylogenetic analysis we detected three and four lineages based on the ORF1 and ORF2 nonrecombinant sequences, respectively. The evolutionary rates of the two BBWV2 ORF coding sequences were 8.895 × 10-4 and 4.560 × 10-4 subs/site/year, respectively. We found a relatively conserved and stable genomic composition with a lower codon usage choice in the two BBWV2 protein coding sequences. ENC-plot and neutrality plot analyses showed that natural selection is the key factor shaping the codon usage pattern of BBWV2. Strong correlations between BBWV2 and broad bean and pepper were observed from similarity index (SiD), codon adaptation index (CAI) and relative codon deoptimization index (RCDI) analyses. Our study is the first to evaluate the phylodynamics, codon usage patterns and adaptive evolution of a fabavirus, and our results may be useful for the understanding of the origin of this virus.
Keywords: broad bean wilt virus 2; codon usage pattern; host adaptation; natural selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Comprehensive codon usage analysis of rice black-streaked dwarf virus based on P8 and P10 protein coding sequences.Infect Genet Evol. 2020 Dec;86:104601. doi: 10.1016/j.meegid.2020.104601. Epub 2020 Oct 27. Infect Genet Evol. 2020. PMID: 33122052
-
A determinant of disease symptom severity is located in RNA2 of broad bean wilt virus 2.Virus Res. 2016 Jan 4;211:25-8. doi: 10.1016/j.virusres.2015.09.018. Epub 2015 Sep 30. Virus Res. 2016. PMID: 26428303
-
Genetic changes and host adaptability in sugarcane mosaic virus based on complete genome sequences.Mol Phylogenet Evol. 2020 Aug;149:106848. doi: 10.1016/j.ympev.2020.106848. Epub 2020 May 5. Mol Phylogenet Evol. 2020. PMID: 32380283
-
Movement protein of broad bean wilt virus 2 serves as a determinant of symptom severity in pepper.Virus Res. 2017 Oct 15;242:141-145. doi: 10.1016/j.virusres.2017.09.024. Epub 2017 Sep 29. Virus Res. 2017. PMID: 28970056
-
Evolution and host adaptability of plant RNA viruses: Research insights on compositional biases.Comput Struct Biotechnol J. 2022 May 17;20:2600-2610. doi: 10.1016/j.csbj.2022.05.021. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35685354 Free PMC article. Review.
Cited by
-
Host Plants Shape the Codon Usage Pattern of Turnip Mosaic Virus.Viruses. 2022 Oct 15;14(10):2267. doi: 10.3390/v14102267. Viruses. 2022. PMID: 36298822 Free PMC article.
-
Identification of a Broad Bean Wilt Virus 2 (BBWV2) Isolate (BBWV2-SP) from Spinacia oleracea L.Int J Mol Sci. 2025 Jun 20;26(13):5946. doi: 10.3390/ijms26135946. Int J Mol Sci. 2025. PMID: 40649722 Free PMC article.
-
Synonymous Codon Usage Analysis of Three Narcissus Potyviruses.Viruses. 2022 Apr 19;14(5):846. doi: 10.3390/v14050846. Viruses. 2022. PMID: 35632588 Free PMC article.
-
Molecular Characterization and Genetic Diversity of Broad Bean Wilt Virus 2 (BBWV-2) from.ACS Omega. 2025 Jul 17;10(29):32433-32443. doi: 10.1021/acsomega.5c04600. eCollection 2025 Jul 29. ACS Omega. 2025. PMID: 40757346 Free PMC article.
-
An Evolutionary Perspective of Codon Usage Pattern, Dinucleotide Composition and Codon Pair Bias in Prunus Necrotic Ringspot Virus.Genes (Basel). 2023 Aug 28;14(9):1712. doi: 10.3390/genes14091712. Genes (Basel). 2023. PMID: 37761852 Free PMC article.
References
-
- Castrovilli S., Savino V., Castellano M.A., Engelbrecht D.J. Characterization of a grapevine isolate of Broad bean wilt virus. Phytopathol. Mediterr. 1985;24:35–40.
-
- Fortass M., Bos L. Survey of faba bean (Vicia faba L.) for viruses in Morocco. Neth. J. Plant. Pathol. 1991;97:369–380. doi: 10.1007/BF03041385. - DOI
-
- Kondo T., Fuji S., Yamashita K., Kang D.K., Chang M.U. Broad bean wilt virus 2 in yams. J. Gen. Plant. Pathol. 2005;71:441–443. doi: 10.1007/s10327-005-0236-x. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

