Cryo-soft X-ray tomography: using soft X-rays to explore the ultrastructure of whole cells
- PMID: 33525785
- PMCID: PMC7289011
- DOI: 10.1042/ETLS20170086
Cryo-soft X-ray tomography: using soft X-rays to explore the ultrastructure of whole cells
Abstract
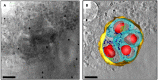
Cryo-soft X-ray tomography is an imaging technique that addresses the need for mesoscale imaging of cellular ultrastructure of relatively thick samples without the need for staining or chemical modification. It allows the imaging of cellular ultrastructure to a resolution of 25-40 nm and can be used in correlation with other imaging modalities, such as electron tomography and fluorescence microscopy, to further enhance the information content derived from biological samples. An overview of the technique, discussion of sample suitability and information about sample preparation, data collection and data analysis is presented here. Recent developments and future outlook are also discussed.
© 2018 The Author(s).
Conflict of interest statement
The Authors declare that there are no competing interests with the manuscript.
Figures




References
-
- Pawley, J.B. (2006) Handbook of Biological Confocal Microscopy. Springer; 985 p
LinkOut - more resources
Full Text Sources
