EPHA7 mutation as a predictive biomarker for immune checkpoint inhibitors in multiple cancers
- PMID: 33526018
- PMCID: PMC7852135
- DOI: 10.1186/s12916-020-01899-x
EPHA7 mutation as a predictive biomarker for immune checkpoint inhibitors in multiple cancers
Abstract
Background: A critical and challenging process in immunotherapy is to identify cancer patients who could benefit from immune checkpoint inhibitors (ICIs). Exploration of predictive biomarkers could help to maximize the clinical benefits. Eph receptors have been shown to play essential roles in tumor immunity. However, the association between EPH gene mutation and ICI response is lacking.
Methods: Clinical data and whole-exome sequencing (WES) data from published studies were collected and consolidated as a discovery cohort to analyze the association between EPH gene mutation and efficacy of ICI therapy. Another independent cohort from Memorial Sloan Kettering Cancer Center (MSKCC) was adopted to validate our findings. The Cancer Genome Atlas (TCGA) cohort was used to perform anti-tumor immunity and pathway enrichment analysis.
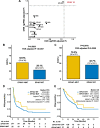
Results: Among fourteen EPH genes, EPHA7-mutant (EPHA7-MUT) was enriched in patients responding to ICI therapy (FDR adjusted P < 0.05). In the discovery cohort (n = 386), significant differences were detected between EPHA7-MUT and EPHA7-wildtype (EPHA7-WT) patients regarding objective response rate (ORR, 52.6% vs 29.1%, FDR adjusted P = 0.0357) and durable clinical benefit (DCB, 70.3% vs 42.7%, FDR adjusted P = 0.0200). In the validation cohort (n = 1144), significant overall survival advantage was observed in EPHA7-MUT patients (HR = 0.62 [95% confidence interval, 0.39 to 0.97], multivariable adjusted P = 0.0367), which was independent of tumor mutational burden (TMB) and copy number alteration (CNA). Notably, EPHA7-MUT patients without ICI therapy had significantly worse overall survival in TCGA cohort (HR = 1.33 [95% confidence interval, 1.06 to 1.67], multivariable adjusted P = 0.0139). Further gene set enrichment analysis revealed enhanced anti-tumor immunity in EPHA7-MUT tumor.
Conclusions: EPHA7-MUT successfully predicted better clinical outcomes in ICI-treated patients across multiple cancer types, indicating that EPHA7-MUT could serve as a potential predictive biomarker for immune checkpoint inhibitors.
Keywords: Biomarker; EPHA7; Eph receptors; Immune checkpoint inhibitor; Pan-cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Gutzmer R, Stroyakovskiy D, Gogas H, Robert C, Lewis K, Protsenko S, et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAFV600 mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;395:1835–1844. doi: 10.1016/S0140-6736(20)30934-X. - DOI - PubMed
-
- Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, de Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/ mismatch repair–deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

