Induction of ADAM10 by Radiation Therapy Drives Fibrosis, Resistance, and Epithelial-to-Mesenchyal Transition in Pancreatic Cancer
- PMID: 33526513
- PMCID: PMC8260469
- DOI: 10.1158/0008-5472.CAN-20-3892
Induction of ADAM10 by Radiation Therapy Drives Fibrosis, Resistance, and Epithelial-to-Mesenchyal Transition in Pancreatic Cancer
Abstract
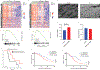
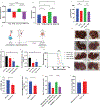
Stromal fibrosis activates prosurvival and proepithelial-to-mesenchymal transition (EMT) pathways in pancreatic ductal adenocarcinoma (PDAC). In patient tumors treated with neoadjuvant stereotactic body radiation therapy (SBRT), we found upregulation of fibrosis, extracellular matrix (ECM), and EMT gene signatures, which can drive therapeutic resistance and tumor invasion. Molecular, functional, and translational analysis identified two cell-surface proteins, a disintegrin and metalloprotease 10 (ADAM10) and ephrinB2, as drivers of fibrosis and tumor progression after radiation therapy (RT). RT resulted in increased ADAM10 expression in tumor cells, leading to cleavage of ephrinB2, which was also detected in plasma. Pharmacologic or genetic targeting of ADAM10 decreased RT-induced fibrosis and tissue tension, tumor cell migration, and invasion, sensitizing orthotopic tumors to radiation killing and prolonging mouse survival. Inhibition of ADAM10 and genetic ablation of ephrinB2 in fibroblasts reduced the metastatic potential of tumor cells after RT. Stimulation of tumor cells with ephrinB2 FC protein reversed the reduction in tumor cell invasion with ADAM10 ablation. These findings represent a model of PDAC adaptation that explains resistance and metastasis after RT and identifies a targetable pathway to enhance RT efficacy. SIGNIFICANCE: Targeting a previously unidentified adaptive resistance mechanism to radiation therapy in PDAC tumors in combination with radiation therapy could increase survival of the 40% of PDAC patients with locally advanced disease.See related commentary by Garcia Garcia et al., p. 3158 GRAPHICAL ABSTRACT: http://cancerres.aacrjournals.org/content/canres/81/12/3255/F1.large.jpg.
©2021 American Association for Cancer Research.
Conflict of interest statement
Conflict of Interest Statements
Dr. Karam receives clinical trial research funding unrelated to this work from AstraZeneca.
Figures




Comment in
-
ADAM10 Evens Out the Double-Edged Sword of Radiotherapy in Pancreatic Cancer.Cancer Res. 2021 Jun 15;81(12):3158-3159. doi: 10.1158/0008-5472.CAN-21-0519. Cancer Res. 2021. PMID: 34224375
Similar articles
-
ADAM10 Evens Out the Double-Edged Sword of Radiotherapy in Pancreatic Cancer.Cancer Res. 2021 Jun 15;81(12):3158-3159. doi: 10.1158/0008-5472.CAN-21-0519. Cancer Res. 2021. PMID: 34224375
-
Aspartate β-hydroxylase promotes pancreatic ductal adenocarcinoma metastasis through activation of SRC signaling pathway.J Hematol Oncol. 2019 Dec 30;12(1):144. doi: 10.1186/s13045-019-0837-z. J Hematol Oncol. 2019. PMID: 31888763 Free PMC article.
-
Trop-2 induces ADAM10-mediated cleavage of E-cadherin and drives EMT-less metastasis in colon cancer.Neoplasia. 2021 Sep;23(9):898-911. doi: 10.1016/j.neo.2021.07.002. Epub 2021 Jul 25. Neoplasia. 2021. PMID: 34320447 Free PMC article.
-
Targeting ADAM10 in Renal Diseases.Curr Mol Med. 2023;23(10):1037-1045. doi: 10.2174/1566524023666221020142504. Curr Mol Med. 2023. PMID: 36278450 Review.
-
Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins.Biochem Soc Trans. 2017 Aug 15;45(4):937-44. doi: 10.1042/BST20160296. Epub 2017 Jul 7. Biochem Soc Trans. 2017. PMID: 28687716 Review.
Cited by
-
High local failure rates despite high margin-negative resection rates in a cohort of borderline resectable and locally advanced pancreatic cancer patients treated with stereotactic body radiation therapy following multi-agent chemotherapy.Cancer Med. 2022 Apr;11(7):1659-1668. doi: 10.1002/cam4.4527. Epub 2022 Feb 10. Cancer Med. 2022. PMID: 35142085 Free PMC article.
-
Sequence basis for selectivity of ephrin-B2 ligand for Eph receptors and pathogenic henipavirus G glycoproteins.J Virol. 2023 Nov 30;97(11):e0062123. doi: 10.1128/jvi.00621-23. Epub 2023 Oct 16. J Virol. 2023. PMID: 37931130 Free PMC article.
-
Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma.J Cell Mol Med. 2023 Apr;27(8):1144-1152. doi: 10.1111/jcmm.17728. Epub 2023 Mar 22. J Cell Mol Med. 2023. PMID: 36946281 Free PMC article.
-
Simultaneous targeting of PD-1 and IL-2Rβγ with radiation therapy inhibits pancreatic cancer growth and metastasis.Cancer Cell. 2023 May 8;41(5):950-969.e6. doi: 10.1016/j.ccell.2023.04.001. Epub 2023 Apr 27. Cancer Cell. 2023. PMID: 37116489 Free PMC article.
-
The Cellular Origins of Cancer-Associated Fibroblasts and Their Opposing Contributions to Pancreatic Cancer Growth.Front Cell Dev Biol. 2021 Sep 27;9:743907. doi: 10.3389/fcell.2021.743907. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34646829 Free PMC article. Review.
References
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

