Holin-Dependent Secretion of the Large Clostridial Toxin TpeL by Clostridium perfringens
- PMID: 33526612
- PMCID: PMC8088506
- DOI: 10.1128/JB.00580-20
Holin-Dependent Secretion of the Large Clostridial Toxin TpeL by Clostridium perfringens
Abstract
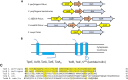
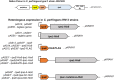
Large clostridial toxins (LCTs) are secreted virulence factors found in several species, including Clostridioides difficile, Clostridium perfringens, Paeniclostridium sordellii, and Clostridium novyi LCTs are large toxins that lack a secretion signal sequence, and studies by others have shown that the LCTs of C. difficile, TcdA and TcdB, require a holin-like protein, TcdE, for secretion. The TcdE gene is located on the pathogenicity locus (PaLoc) of C. difficile, and holin-encoding genes are also present in the LCT-encoded PaLocs from P. sordellii and C. perfringens However, the holin (TpeE) associated with the C. perfringens LCT TpeL has no homology and a different membrane topology than TcdE. In addition, TpeE has a membrane topology identical to that of the TatA protein, which is the core of the twin-arginine translocation (Tat) secretion system. To determine if TpeE was necessary and sufficient to secrete TpeL, the genes from a type C strain of C. perfringens were expressed in a type A strain of C. perfringens, HN13, and secretion was measured using Western blot methods. We found that TpeE was required for TpeL secretion and that secretion was not due to cell lysis. Mutant forms of TpeE lacking an amphipathic helix and a charged C-terminal domain failed to secrete TpeL, and mutations that deleted conserved LCT domains in TpeL indicated that only the full-length protein could be secreted. In summary, we have identified a novel family of holin-like proteins that can function, in some cases, as a system of protein secretion for proteins that need to fold in the cytoplasm.IMPORTANCE Little is known about the mechanism by which LCTs are secreted. Since LCTs are major virulence factors in clostridial pathogens, we wanted to define the mechanism by which an LCT in C. perfringens, TpeL, is secreted by a protein (TpeE) lacking homology to previously described secretion-associated holins. We discovered that TpeE is a member of a widely dispersed class of holin proteins, and TpeE is necessary for the secretion of TpeL. TpeE bears a high degree of similarity in membrane topology to TatA proteins, which form the pore through which Tat secretion substrates pass through the cytoplasmic membrane. Thus, the TpeE-TpeL secretion system may be a model for understanding not only holin-dependent secretion but also how TatA proteins function in the secretion process.
Keywords: Clostridium; molecular genetics; perfringens; secretion systems; toxin.
Copyright © 2021 American Society for Microbiology.
Figures





Similar articles
-
A Highly Specific Holin-Mediated Mechanism Facilitates the Secretion of Lethal Toxin TcsL in Paeniclostridiumsordellii.Toxins (Basel). 2022 Feb 8;14(2):124. doi: 10.3390/toxins14020124. Toxins (Basel). 2022. PMID: 35202151 Free PMC article.
-
Expression of the large clostridial toxins is controlled by conserved regulatory mechanisms.Int J Med Microbiol. 2014 Nov;304(8):1147-59. doi: 10.1016/j.ijmm.2014.08.008. Epub 2014 Aug 18. Int J Med Microbiol. 2014. PMID: 25190355
-
Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE.PLoS Pathog. 2012;8(6):e1002727. doi: 10.1371/journal.ppat.1002727. Epub 2012 Jun 7. PLoS Pathog. 2012. PMID: 22685398 Free PMC article.
-
Large Clostridial Toxins: Mechanisms and Roles in Disease.Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0006421. doi: 10.1128/MMBR.00064-21. Epub 2021 Jun 2. Microbiol Mol Biol Rev. 2021. PMID: 34076506 Free PMC article. Review.
-
Occurrence and potential mechanism of holin-mediated non-lytic protein translocation in bacteria.Microb Cell. 2022 Sep 23;9(10):159-173. doi: 10.15698/mic2022.10.785. eCollection 2022 Oct 3. Microb Cell. 2022. PMID: 36262927 Free PMC article. Review.
Cited by
-
Type IV Pili Are a Critical Virulence Factor in Clinical Isolates of Paenibacillus thiaminolyticus.mBio. 2022 Dec 20;13(6):e0268822. doi: 10.1128/mbio.02688-22. Epub 2022 Nov 14. mBio. 2022. PMID: 36374038 Free PMC article.
-
A sequence invariable region in TcdB2 is required for toxin escape from Clostridioides difficile.J Bacteriol. 2024 Jul 25;206(7):e0009624. doi: 10.1128/jb.00096-24. Epub 2024 Jun 18. J Bacteriol. 2024. PMID: 38888328 Free PMC article.
-
PncsHub: a platform for annotating and analyzing non-classically secreted proteins in Gram-positive bacteria.Nucleic Acids Res. 2022 Jan 7;50(D1):D848-D857. doi: 10.1093/nar/gkab814. Nucleic Acids Res. 2022. PMID: 34551435 Free PMC article.
-
Yersinia entomophaga Tc toxin is released by T10SS-dependent lysis of specialized cell subpopulations.Nat Microbiol. 2024 Feb;9(2):390-404. doi: 10.1038/s41564-023-01571-z. Epub 2024 Jan 18. Nat Microbiol. 2024. PMID: 38238469 Free PMC article.
-
A Clostridioides difficile endolysin modulates toxin secretion without cell lysis.Commun Biol. 2024 Aug 23;7(1):1044. doi: 10.1038/s42003-024-06730-4. Commun Biol. 2024. PMID: 39179651 Free PMC article.
References
-
- Guttenberg G, Hornei S, Jank T, Schwan C, Lu W, Einsle O, Papatheodorou P, Aktories K. 2012. Molecular characteristics of Clostridium perfringens TpeL toxin and consequences of mono-O-GlcNAcylation of Ras in living cells. J Biol Chem 287:24929–24940. doi:10.1074/jbc.M112.347773. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

