Consequences of aneuploidy in human fibroblasts with trisomy 21
- PMID: 33526671
- PMCID: PMC8017964
- DOI: 10.1073/pnas.2014723118
Consequences of aneuploidy in human fibroblasts with trisomy 21
Abstract
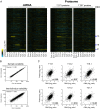
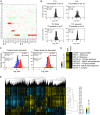
An extra copy of chromosome 21 causes Down syndrome, the most common genetic disease in humans. The mechanisms contributing to aneuploidy-related pathologies in this syndrome, independent of the identity of the triplicated genes, are not well defined. To characterize aneuploidy-driven phenotypes in trisomy 21 cells, we performed global transcriptome, proteome, and phenotypic analyses of primary human fibroblasts from individuals with Patau (trisomy 13), Edwards (trisomy 18), or Down syndromes. On average, mRNA and protein levels were increased by 1.5-fold in all trisomies, with a subset of proteins enriched for subunits of macromolecular complexes showing signs of posttranscriptional regulation. These results support the lack of evidence for widespread dosage compensation or dysregulation of chromosomal domains in human autosomes. Furthermore, we show that several aneuploidy-associated phenotypes are present in trisomy 21 cells, including lower viability and increased dependency on serine-driven lipid synthesis. Our studies establish a critical role of aneuploidy, independent of triplicated gene identity, in driving cellular defects associated with trisomy 21.
Keywords: Down syndrome; aneuploidy; dosage compensation; sphingolipids; trisomy 21.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

