Activation of meiotic recombination by nuclear import of the DNA break hotspot-determining complex in fission yeast
- PMID: 33526714
- PMCID: PMC7929924
- DOI: 10.1242/jcs.253518
Activation of meiotic recombination by nuclear import of the DNA break hotspot-determining complex in fission yeast
Abstract
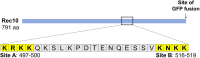
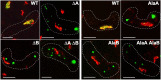
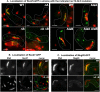
Meiotic recombination forms crossovers important for proper chromosome segregation and offspring viability. This complex process involves many proteins acting at each of the multiple steps of recombination. Recombination initiates by formation of DNA double-strand breaks (DSBs), which in the several species examined occur with high frequency at special sites (DSB hotspots). In Schizosaccharomyces pombe, DSB hotspots are bound with high specificity and strongly activated by linear element (LinE) proteins Rec25, Rec27 and Mug20, which form colocalized nuclear foci with Rec10, essential for all DSB formation and recombination. Here, we test the hypothesis that the nuclear localization signal (NLS) of Rec10 is crucial for coordinated nuclear entry after forming a complex with other LinE proteins. In NLS mutants, all LinE proteins were abundant in the cytoplasm, not the nucleus; DSB formation and recombination were much reduced but not eliminated. Nuclear entry of limited amounts of Rec10, apparently small enough for passive nuclear entry, can account for residual recombination. LinE proteins are related to synaptonemal complex proteins of other species, suggesting that they also share an NLS, not yet identified, and undergo protein complex formation before nuclear entry.This article has an associated First Person interview with Mélody Wintrebert, joint first author of the paper.
Keywords: DSB hotspot determinants; Linear element proteins; Meiotic recombination; Nuclear localization signal; S. pombe; Synaptonemal complex proteins.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



Similar articles
-
Functional organization of protein determinants of meiotic DNA break hotspots.Sci Rep. 2017 May 3;7(1):1393. doi: 10.1038/s41598-017-00742-3. Sci Rep. 2017. PMID: 28469148 Free PMC article.
-
Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination.Curr Biol. 2008 Jun 3;18(11):849-54. doi: 10.1016/j.cub.2008.05.025. Curr Biol. 2008. PMID: 18514516 Free PMC article.
-
Protein determinants of meiotic DNA break hot spots.Mol Cell. 2013 Mar 7;49(5):983-96. doi: 10.1016/j.molcel.2013.01.008. Epub 2013 Feb 7. Mol Cell. 2013. PMID: 23395004 Free PMC article.
-
The conserved histone variant H2A.Z illuminates meiotic recombination initiation.Curr Genet. 2018 Oct;64(5):1015-1019. doi: 10.1007/s00294-018-0825-9. Epub 2018 Mar 16. Curr Genet. 2018. PMID: 29549582 Review.
-
Regulation Mechanisms of Meiotic Recombination Revealed from the Analysis of a Fission Yeast Recombination Hotspot ade6-M26.Biomolecules. 2022 Nov 26;12(12):1761. doi: 10.3390/biom12121761. Biomolecules. 2022. PMID: 36551189 Free PMC article. Review.
Cited by
-
Mug20-Rec25-Rec27 binds DNA and enhances meiotic DNA break formation via phase-separated condensates.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf123. doi: 10.1093/nar/gkaf123. Nucleic Acids Res. 2025. PMID: 40037704 Free PMC article.
-
Dynamic configurations of meiotic DNA-break hotspot determinant proteins.J Cell Sci. 2022 Feb 1;135(3):jcs259061. doi: 10.1242/jcs.259061. Epub 2022 Feb 7. J Cell Sci. 2022. PMID: 35028663 Free PMC article.
-
Redirecting meiotic DNA break hotspot determinant proteins alters localized spatial control of DNA break formation and repair.Nucleic Acids Res. 2022 Jan 25;50(2):899-914. doi: 10.1093/nar/gkab1253. Nucleic Acids Res. 2022. PMID: 34967417 Free PMC article.
-
Beyond microtubules: The cellular environment at the endoplasmic reticulum attracts proteins to the nucleus, enabling nuclear transport.iScience. 2024 Feb 15;27(3):109235. doi: 10.1016/j.isci.2024.109235. eCollection 2024 Mar 15. iScience. 2024. PMID: 38439967 Free PMC article.
References
-
- Bähler, J., Wu, J.-Q., Longtine, M. S., Shah, N. G., McKenzie, A., III, Steever, A. B., Wach, A., Philippsen, P. and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

