Genome sequencing broadens the range of contributing variants with clinical implications in schizophrenia
- PMID: 33526774
- PMCID: PMC7851385
- DOI: 10.1038/s41398-021-01211-2
Genome sequencing broadens the range of contributing variants with clinical implications in schizophrenia
Abstract
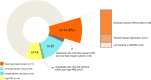
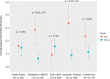
The range of genetic variation with potential clinical implications in schizophrenia, beyond rare copy number variants (CNVs), remains uncertain. We therefore analyzed genome sequencing data for 259 unrelated adults with schizophrenia from a well-characterized community-based cohort previously examined with chromosomal microarray for CNVs (none with 22q11.2 deletions). We analyzed these genomes for rare high-impact variants considered causal for neurodevelopmental disorders, including single-nucleotide variants (SNVs) and small insertions/deletions (indels), for potential clinical relevance based on findings for neurodevelopmental disorders. Also, we investigated a novel variant type, tandem repeat expansions (TREs), in 45 loci known to be associated with monogenic neurological diseases. We found several of these variants in this schizophrenia population suggesting that these variants have a wider clinical spectrum than previously thought. In addition to known pathogenic CNVs, we identified 11 (4.3%) individuals with clinically relevant SNVs/indels in genes converging on schizophrenia-relevant pathways. Clinical yield was significantly enriched in females and in those with broadly defined learning/intellectual disabilities. Genome analyses also identified variants with potential clinical implications, including TREs (one in DMPK; two in ATXN8OS) and ultra-rare loss-of-function SNVs in ZMYM2 (a novel candidate gene for schizophrenia). Of the 233 individuals with no pathogenic CNVs, we identified rare high-impact variants (i.e., clinically relevant or with potential clinical implications) for 14 individuals (6.0%); some had multiple rare high-impact variants. Mean schizophrenia polygenic risk score was similar between individuals with and without clinically relevant rare genetic variation; common variants were not sufficient for clinical application. These findings broaden the individual and global picture of clinically relevant genetic risk in schizophrenia, and suggest the potential translational value of genome sequencing as a single genetic technology for schizophrenia.
Conflict of interest statement
Daniele Merico is a full-time employee of Deep Genomics Inc. and is entitled to a stock option. All other authors report no financial relationships with commercial interests.
Figures
References
-
- Cannon TD, Kaprio J, Lönnqvist J, Huttunen M, Koskenvuo M. The genetic epidemiology of schizophrenia in a Finnish twin cohort: a population-based modeling study. JAMA Psychiatry. 1998;55:67–74. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

