CNS fibroblasts form a fibrotic scar in response to immune cell infiltration
- PMID: 33526922
- PMCID: PMC7877789
- DOI: 10.1038/s41593-020-00770-9
CNS fibroblasts form a fibrotic scar in response to immune cell infiltration
Abstract
Fibrosis is a common pathological response to inflammation in many peripheral tissues and can prevent tissue regeneration and repair. Here, we identified persistent fibrotic scarring in the CNS following immune cell infiltration in the experimental autoimmune encephalomyelitis (EAE) mouse model of multiple sclerosis. Using lineage tracing and single-cell sequencing in EAE, we determined that the majority of the fibrotic scar is derived from proliferative CNS fibroblasts, not pericytes or infiltrating bone marrow-derived cells. Ablating proliferating fibrotic cells using cell-specific expression of herpes thymidine kinase led to an increase in oligodendrocyte lineage cells within the inflammatory lesions and a reduction in motor disability. We further identified that interferon-gamma pathway genes are enriched in CNS fibrotic cells, and the fibrotic cell-specific deletion of Ifngr1 resulted in reduced fibrotic scarring in EAE. These data delineate a framework for understanding the CNS fibrotic response.
Conflict of interest statement
Figures
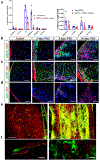
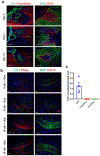

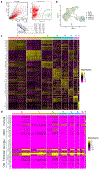
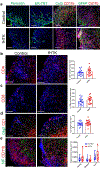
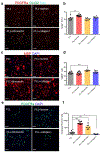

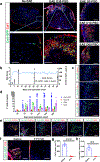
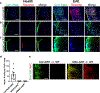

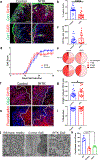

Comment in
-
Inflammation drives fibrotic scars in the CNS.Nat Neurosci. 2021 Feb;24(2):157-159. doi: 10.1038/s41593-020-00777-2. Nat Neurosci. 2021. PMID: 33526921 No abstract available.
References
Method only References
-
- Wendling O, Bornert JM, Chambon P & Metzger D Efcient temporally controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47, 14–18 (2009). - PubMed
-
- Heng TSP et al. Te Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol 9, 1091–1094 (2008). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

