Metabolic control of DNA methylation in naive pluripotent cells
- PMID: 33526924
- PMCID: PMC7116828
- DOI: 10.1038/s41588-020-00770-2
Metabolic control of DNA methylation in naive pluripotent cells
Abstract
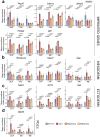
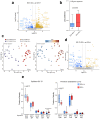
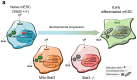
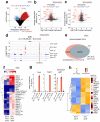
Naive epiblast and embryonic stem cells (ESCs) give rise to all cells of adults. Such developmental plasticity is associated with genome hypomethylation. Here, we show that LIF-Stat3 signaling induces genomic hypomethylation via metabolic reconfiguration. Stat3-/- ESCs show decreased α-ketoglutarate production from glutamine, leading to increased Dnmt3a and Dnmt3b expression and DNA methylation. Notably, genome methylation is dynamically controlled through modulation of α-ketoglutarate availability or Stat3 activation in mitochondria. Alpha-ketoglutarate links metabolism to the epigenome by reducing the expression of Otx2 and its targets Dnmt3a and Dnmt3b. Genetic inactivation of Otx2 or Dnmt3a and Dnmt3b results in genomic hypomethylation even in the absence of active LIF-Stat3. Stat3-/- ESCs show increased methylation at imprinting control regions and altered expression of cognate transcripts. Single-cell analyses of Stat3-/- embryos confirmed the dysregulated expression of Otx2, Dnmt3a and Dnmt3b as well as imprinted genes. Several cancers display Stat3 overactivation and abnormal DNA methylation; therefore, the molecular module that we describe might be exploited under pathological conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures
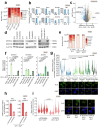

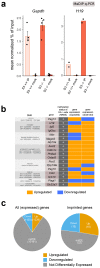
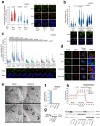
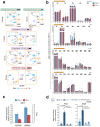


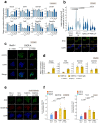
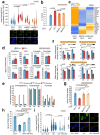

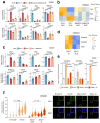

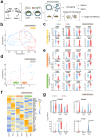
References
-
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Dev Camb Engl. 1987;99:371–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

