Gene-based therapies for neurodegenerative diseases
- PMID: 33526943
- PMCID: PMC8394447
- DOI: 10.1038/s41593-020-00778-1
Gene-based therapies for neurodegenerative diseases
Abstract
Gene therapy is making a comeback. With its twin promise of targeting disease etiology and 'long-term correction', gene-based therapies (defined here as all forms of genome manipulation) are particularly appealing for neurodegenerative diseases, for which conventional pharmacologic approaches have been largely disappointing. The recent success of a viral-vector-based gene therapy in spinal muscular atrophy-promoting survival and motor function with a single intravenous injection-offers a paradigm for such therapeutic intervention and a platform to build on. Although challenges remain, the newfound optimism largely stems from advances in the development of viral vectors that can diffusely deliver genes throughout the CNS, as well as genome-engineering tools that can manipulate disease pathways in ways that were previously impossible. Surely spinal muscular atrophy cannot be the only neurodegenerative disease amenable to gene therapy, and one can imagine a future in which the toolkit of a clinician will include gene-based therapeutics. The goal of this Review is to highlight advances in the development and application of gene-based therapies for neurodegenerative diseases and offer a prospective look into this emerging arena.
Conflict of interest statement
Figures
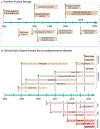

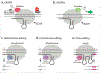
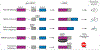

References
-
- Keeler CE Gene therapy. J Hered 38, 294–298 (1947). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

